Systemic lupus erythematosus (SLE) is an autoimmune disease in which the immune system abnormally reacts against cells and tissues leading to inflammation. Epigenetic alterations, including DNA methylation and histone modification, have critical effects on autoimmune disease and SLE pathogenesis via dysregulation of critical genes.
AimsThe purpose of this study was to evaluate the epigenetic-related gene expression of DNA methyltransferase (DNMT) and histone deacetylase 1 (HDAC1) in Iranian patients with SLE.
MethodsThis matched case–control study included 16 people with SLE and 16 healthy people who were referred to the Rafsanjani rheumatology clinic, in southeast Iran. The expression of DNMT and HDAC1 genes was measured through a real-time PCR assay of blood samples.
ResultsDNMT gene expression did not differ significantly between SLE and healthy groups (P=0.21). In contrast, HDAC1 gene expression was enhanced in the SLE group, but this enhancement failed to reach statistical significance (P=0.94).
ConclusionThe results of this study suggest that overexpression of HDAC1 could serve as a diagnostic for SLE disease. Additional studies with larger sample sizes are required to confirm our findings. Evaluation of other genes related to SLE disease is essential and may help to make an accurate diagnosis of the disease.
El lupus eritematoso sistémico (LES) es una enfermedad autoinmune, en la cual el sistema inmunitario reacciona de manera anormal frente a las células y tejidos causantes de la inflamación. Las alteraciones epigenéticas, incluyendo la metilación del ADN y la modificación de la histona, tienen efectos críticos en la enfermedad autoinmune y la patogenia del LES, a través de la desregulación de los genes críticos.
ObjetivoEl objetivo de este estudio fue evaluar la expresión del gen relacionado con la epigenética de ADN metiltransferasa (DNMT) e histona deacetilasa 1 (HDAC1) en los pacientes iraníes afectados de LES.
MétodosEste estudio pareado caso-control incluyó 16 personas con LES y 16 personas sanas, derivadas a la clínica de reumatología de Rafsanjan, en el sudeste de Irán. La expresión de los genes DNMT y HDAC1 se midió mediante una PCR a tiempo real de muestras de sangre.
ResultadosLa expresión del gen DNMT no difirió significativamente entre los grupos de pacientes de LES y de controles sanos (p=0,21). Por contra, la expresión del gen HDAC1 se incrementó en el grupo LES, aunque dicho incremento no alcanzó significación estadística (p=0,94).
ConclusiónLos resultados de este estudio sugieren que la sobreexpresión de HDAC1 podría servir para diagnosticar el LES. Son necesarios estudios adicionales con muestras de mayor tamaño para confirmar nuestros hallazgos. Es esencial la evaluación de otros genes relacionados con el LES, pudiendo ayudar a realizar un diagnóstico preciso de la enfermedad.
Systemic lupus erythematosus (SLE) is an autoimmune disease that is associated with vascular and connective tissue inflammation. SLE affects a variety of organs including the joints, skin, kidneys, and heart.1 SLE can also cause significant problems in the nervous system. SLE occurs all over the world, with a prevalence of 20–150 cases per 100,000 people. In Iran, SLE occurs in about 40 cases per 100,000 people.2,3
SLE may affect active B cells and T cells causing abnormal differentiation. Increased activation of these cells leads to the production of antibodies against nucleic acids, proteins, and ribonucleoproteins. Clinical symptoms vary based on the type of damaged tissue. SLE is also determined by hereditary and environmental factors including ultraviolet radiation and drugs.4,5
SLE causes extensive damage to the connective tissue, blood vessels, and serous membranes.6 The progression of SLE is unpredictable, involves many changes that lead to progressive disability in young patients, and has a variety of harmful effects on physical, psychological, and social health fields.7,8
Epigenetics is the study of the heritable changes in the function and expression of genes, in the absence of changes in the DNA sequence. Epigenetic mechanisms include DNA methylation, histone modification, and alteration in transcription factors9 that lead to the expression or non-expression of genes.10 Epigenetic changes can be determined by evaluating DNA methyltransferase (DNMT) and histone deacetylase 1 (HDAC1), enzymes related to DNA methylation and acetylation. The reduction of DNMT1 expression and enhancement of methylation-sensitive autoimmune genes activation in T cells of patients with SLE could be a part of epigenetic changes.11 The relationship between DNMT1 and HDAC1 and SLE and other autoimmune diseases has been reported,11 suggesting the study of epigenetic mechanisms in regulating gene expression and the effect of drugs on these genes. Different environmental pollutions can lead to epigenetic changes,12 and that, in turn, may cause autoimmune diseases such as SLE.13 Rafsanjan city in the southeast of Iran is prone to environmental pollution, including agricultural pesticides and contaminants from Sarcheshmeh copper mine pollutants which may contribute to epigenetic changes. Therefore, the evaluation of related epigenetic genes in patients in Rafsanjan with SLE is warranted.
In this study, we examined the DNMT and HDAC1 genes expression in Iranian SLE patients referred to the Rafsanjan rheumatology clinic.
Materials and methodsStudy setting and participantsA matched case–control study design was used. During three month (1 Oct to 30 Dec 2020), sixteen Iranian patients with SLE admitted to rheumatology clinic in Rafsanjan city, the southeast of Iran, were included in the case group in a consecutive manner. For the control group, sixteen healthy people from the hospital staff of Rafsanjan city selected by random method.
Inclusion and exclusion criteria for case groupThe patients fulfilled the American College of Rheumatology diagnostic criteria for SLE.14 Then 16 patients were appraised with clinical examination (there are no more patients with SLE in Rafsanjan city), and laboratory tests such as ESR, CRP, RF, anti CCP, ANA, anti DS DNA were performed. A rheumatologist then confirmed the results. The voluntary and informed participation in the case group were considered.
Inclusion and exclusion criteria for control groupSixteen adult healthy people (18–65 years old) who had no ACR criteria were recruited from the hospital staff of Rafsanjan. Subjects that had used anti-inflammatory drugs in the last three months or had the main symptoms of SLE in their family were excluded from the control group. The voluntary and informed participation in the control group were considered.
Samples sizeAccording to the inclusion and exclusion criteria, all adult SLE patients (18–65 years old) in Rafsanjan city during 1 Oct to 30 Dec 2020 were enrolled as the case group. The ratio of control to case was 1:1, and matching age and sex.
Collecting dataDemographic and epidemiologic data including age, sex, academic education (BSc, MSc, Ph.D.), smoking at least one cigarette a day, body mass index (BMI), and job status were matched between the two groups.
Experimental procedureA 10ml blood sample was obtained from each subject in both groups. A sample of the blood (3ml) was reserved for ELISA assay, and an additional sample (7ml) was collected in EDTA tubes for the real-time PCR method. Clotted blood was centrifuged for 3–5min with 3000rpm to separate the serum. The serum was kept at −20°C until analyzed for ANA and CCP via ELSA kits (Germany, AESKU) according to the kit protocol.
An RNA extraction kit was applied to extract total RNA from peripheral blood samples. Extracted RNA quality was determined via electrophoresis on the ethidium bromide pretreated agarose gels. Absorption was measured at 260/280nm by a spectrophotometer. cDNA was synthesized using a cDNA synthesis kit (Parstous, Iran) according to the manufacturer's instructions.
5μl SYBR of green master mix (Parstous, Iran), 1μl of the generated cDNA, and 0.8μM of forward and reverse appropriate primers (Table 1) were mixed for real-time quantitative PCR.
The list of the sequence of primers used for real-time PCR in this study.
| Gene | Primer |
|---|---|
| DNA methyltransferase 1 (DNMT1) | Forward: CCGGCCCCGGTTCTT |
| Reverse: GGACCATGGAGCGCTTGA | |
| Histone deacetylase 1 (HDAC1) | Forward: CGCCAAGTGTGTGGAATTTG |
| Reverse: GCCTCCCAGCATCAGCATA | |
| β-Actin (housekeeping gene) | Forward: GATATCGCTGCGCTCGTCG |
| Reverse: CCCATACCCACCATCACACC |
The cycling program on a BIO-RAD CFX96 system (Bio-Rad Company, USA) was completed as follows: one cycle of 94°C for 30s, 45 cycles of 94°C for 5s, and 45 cycles for 34s. This cycle was performed in triplicate, and β-actin was evaluated as a housekeeping gene. 2−ΔΔct formula was applied for the relative amounting of the PCR product. The dissociation stages, melting curves, and quantitative analyses of the data were performed using CFX manager software version 1.1.308.111 (Bio-Rad, USA).
Statistical analysisThe continuous variables were expressed as the mean±SD, and the categorical variables were presented as a percentage and frequency. Because the data showed a non-normal distribution, the Mann–Whitney test was used to compare the parameters between patients and health groups. The relations between parameters were evaluated using the Pearson correlation coefficient. All statistical analyses were performed with SPSS (version 16.0, SPSS Inc, Chicago, IL, USA). A “P-value” less than 0.05 was considered significant.
Ethical considerationsThe study was conducted in accordance with the Declaration of Helsinki. Institutional Review Board approval (code: IR.RUMS.REC.1396.119) was obtained (April 2020). The present study did not interfere with the process of diagnosis and treatment of patients and all participants signed an informed consent form.
ResultsDemographic and epidemiological characteristicsThis case-control study included 16 patients with SLE (case group) and 16 healthy people (control group). Demographic and epidemiological data were matched between two groups (P>0.05) and provided in Table 2. Most subjects in case and control groups were women, married and housekeepers. One person in the control group was a tobacco smoker. The mean age was 43.2±11.4 years and 38.9±12.1 years for case and control groups, respectively (P=0.31). 56.2% and 37.5% of the subjects were more than 41 years old in case and control group, respectively. The highest percentage and frequency for BMI in patients was 25–29.5, with 43.8% that showed most of the patients are overweight. In the control group, healthy BMI (9.5–24.18) was in first order with 45%. The minority of both groups had academic education (BSc, MSc, PhD).
Demographic and epidemiological data in case (16 patients with systemic lupus erythematosus; SLE) and control (16 healthy people) groups.
| Case groupN (%) | Control groupN (%) | P value | |
|---|---|---|---|
| Age (years) | |||
| Up to 30 | 0 | 3 (18.7%) | 0.08 |
| 31–40 | 6 (37.6%) | 7 (43.8%) | |
| 41–50 | 4 (25%) | 5 (31.3%) | |
| 51–60 | 5 (31.2%) | 1 (6.2%) | |
| More than 60 | 1 (6.2%) | 0 | |
| Body mass index (kg/m2) | |||
| Up to 18.5 | 1 (6.2%) | 1 (6.2%) | 0.31 |
| 18.5–24.9 | 3 (18.8%) | 8 (50%) | |
| 25–29.9 | 7 (43.8%) | 6 (37.6%) | |
| 30–34.9 | 4 (25%) | 1 (6.2%) | |
| More than 35 | 1 (6.2%) | 0 (0%) | |
| Gender | |||
| Female | 15 (93.7%) | 14 (87.5%) | 0.55 |
| Male | 1 (6.3%) | 2 (12.5%) | |
| Academic education | |||
| Yes | 2 (12.5%) | 6 (37.6%) | 0.11 |
| No | 14 (87.5%) | 10 (62.5%) | |
| Job status | |||
| Housewife | 15 (93.7%) | 13 (81.2%) | 0.67 |
| Clerk | 1 (6.3%) | 3 (18.8%) | |
| Smoking (one cigarette a day) | |||
| Yes | 0 | 1 (6.3%) | 0.31 |
| No | 16 (100%) | 15 (93.7%) | |
| Marital status | |||
| Married | 14 (87.5%) | 13 (81.2%) | 0.63 |
| Single | 2 (12.5%) | 3 (18.8%) | |
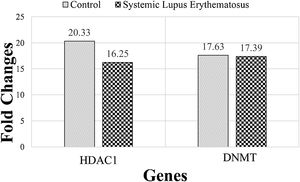
The expression of DNMT and HDAC1 genes were evaluated, via real-time PCR assay, and patients with SLE were compared to healthy group. The mean rank of DNMT gene expression was 17.63 in the SLE group and 17.39 in the control group. Mann–Whitney statistical test reported that the expression of this gene did not differ significantly between SLE and control groups (P=0.21). The mean rank in the expression of the HDAC1 gene was 20.33 and 16.25 in SLE and control groups, respectively. While HDAC1 gene expression was enhanced in the SLE group, this enhancement was not statistically significant (P=0.94) (Fig. 1).
The mean rank of HDAC1 and DNMT genes in systemic lupus erythematosus (SLE) and control groups. There is no significant difference between groups. Results are obtained from three independent experiments and data are presented as mean±SD. The significance level was P˂0.05 (HDAC1: P=0.94), (DNMT: P=0.21).
Comparison of serum levels of the epigenetic genes of 16 patients with SLE and 16 healthy people indicate that the expression of the DNMT gene did not differ between SLE and control groups. While HDAC1 gene expression increased in the SLE group this increase was not significant.
In contrast with our finding, previous studies have evaluated DNA methylation in T cells from SLE patients and found the mean DNMT gene expression significantly diminished.15,16 Pan et al. (2010) also demonstrated that the DNMT gene reduced in patients with SLE.17 Decreasing DNMT gene expression could be the result of inhibition of ERK pathway signaling, which causes overexpression of some genes that improve the SLE disease.18–20 DNMT is reduced in older people and could be a cause of rheumatoid disease.21,22
Consistent with our study, Hu et al., reported that HDAC1 gene expression was not significantly different between patients with SLE and healthy people.23 Nawrocki et al., found HDAC1-3 mRNA expression significantly enhanced in patients with SLE.24 HDAC has been shown to exacerbate inflammation in vitro, and HDAC inhibitors can help in the treatment of inflammation in the arthritis.25 Lin et al., indicated that overexpression of HDAC1 might be a reason for the inflammation, and an HDAC inhibitor could reduce inflammation or disease progression.26 Horiuchi et al., studied the expression of HDAC in rheumatoid arthritis synovial fibroblasts and reported that HDAC1 enhanced synovial fibroblasts.27 Kawabata et al., found that HDAC1 increased in synovial tissue and suggested that the overexpression of HDAC1 might contribute to synovial inflammation.25
In this matched case–control study; most patients were women (93.7%), were over 41 years old (56.2%) and most patients were overweight (BMI=25–29.9kg/m2) (43.8%) and obese (BMI≥30kg/m2) (25%). The majority of patients in our study were women consistent with reports that SLE is more prevalent among females.28 SLE in women has been associated with a number of reproductive factors including oral contraceptive use, older age at menarche (≤10 years) and the adoption of hormone replacement therapy following menopause.29 And sex specific changes in aging B cells that precede autoimmune disease induce have also been identified in mice.30
The population in this study was middle-aged and aging has been associated with a decline in immunity31 that includes changes in autoantibody levels32 that could be influencing the progression of the disease. Obesity has also been shown to contribute to SLE28,33 and rheumatoid arthritis34 – another autoimmune disease – via changes in adipokines, inflammatory cytokines, released from adipose tissue.35,36 Adipose tissue also contains aromatase enzymes and enhances to steroid hormone levels by converting androgens to estrogens.37
Although smoking has been shown to be a risk factor for SLE,38 none of the patients in the present study smoked. Part of this is related to Iranian culture, where smoking is considered to be unflattering to women.
ConclusionIn the present study, although the expression of DNMT was not different between case and control groups, the expression of HDAC1 increased in SLE patients. A larger sample size might support the overexpression of HDAC1 as a diagnostic method for SLE disease as this gene is related to inflammation and rheumatoid disease. Evaluation of other genes that are related to SLE disease is essential and may help an accurate diagnosis of the disease.
Availability of data and materialsThe data used in this study are available from the corresponding author on request.
Ethics approval and consent to participateThe study was conducted in accordance with the Declaration of Helsinki and Institutional Review Board approval has been obtained (IR.RUMS.REC.1396.119).
Consent for publicationBy submitting this document, the authors declare their consent for the final accepted version of the manuscript to be considered for publication.
Authors’ contributionsMA and MRH were responsible for the study concept and design. MA, FM, MMS, and GH led data collection. MA, FM, and MRH were responsible for the analysis and interpretation of data. MA and MM wrote the first draft. JS, MMS, GH, and MRH contributed to the writing of the second and third drafts. JS, MRH, and RH provided comments on initial drafts and coordinated the final draft. All authors read and approved the final manuscript. All authors take responsibility for the integrity of the data and the accuracy of the data analysis.
FundingThanks to financial support, guidance, and advice from the “Rafsanjan University of Medical Sciences”.
Conflicts of interestThe authors of this study declare no conflict of interest.
The authors take this opportunity to thank the Department of Clinical Biochemistry, Faculty of Medicine, Rafsanjan University of Medical Sciences, Rafsanjan, Iran for their financial and technical support.