Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by T-cell immune-dysregulation and loss of tolerance to self-antigens. CTLA-4 and PTPN-22 are involved in the inhibition of T-lymphocytes activation. IL-37 is an anti-inflammatory cytokine that suppresses innate immunity. The relative expression of CTLA-4, IL-37 and PTPN-22 were evaluated as negative regulators of immune response in SLE patients, lupus nephritis (LN) and disease activity.
MethodsReal-Time PCR was performed to determine relative CTLA-4, IL-37, and PTPN-22 mRNA expressions in fifty-eight SLE patients, who were divided into two groups: 29 SLE patients without nephritis and 29 patients with LN, versus fifty controls.
ResultsThere was a significantly high-expression of CTLA-4 and IL-37 genes in SLE patients compared to controls (p=0.005; 0.018 respectively). There was no difference in relative PTPN-22 mRNA expression between the SLE patients and controls. Relative CTLA-4 mRNA expression decreased in LN patients (p=0.044), however, relative IL-37 mRNA over-expressed in LN patients (p=0.001) compared to those without LN. There was a significant over-expression of relative IL-37 andPTPN-22 mRNA in active SLE patients. But, there was a non-significant difference in CTLA-4 expression with disease activity. Regression analysis revealed patients with relative IL-37 mRNA over-expression had two times more to develop lupus nephritis (OR=1.906, 95% CI=1.218–2.983, p=0.005).
ConclusionsRelative IL-37mRNA expression was elevated in SLE patients and associated with renal involvement and disease activity. It could be considered as a new promising predicting tool for LN. Relative PTPN-22 mRNA expression was correlated with disease activity only in SLE patients.
El lupus eritematoso sistémico (LES) es una enfermedad autoinmune caracterizada por la desregulación inmune de las células T, y la pérdida de tolerancia a los antígenos propios. CTLA-4 y PTPN-22 están involucrados en la inhibición de la activación de los linfocitos T. IL-37 es una citocina antiinflamatoria que suprime la inmunidad innata. La expresión relativa de CTLA-4, IL-37 y PTPN-22 se evaluó como reguladores negativos de la respuesta inmune en pacientes con LES, nefritis lúpica (NL) y actividad de la enfermedad.
MétodosSe realizó PCR en tiempo real para determinar las expresiones relativas de ARNm de CTLA-4, IL-37 y PTPN-22 en 58 pacientes con LES, que se dividieron en 2 grupos: 29 pacientes con LES sin nefritis y 29 pacientes con NL frente a 50 controles.
ResultadosHubo una expresión significativamente alta de los genes CTLA-4 e IL-37 en pacientes con LES en comparación con los controles (p=0,005; 0,018, respectivamente). No hubo diferencia en la expresión relativa del ARNm de PTPN-22 entre los pacientes con LES y los controles. La expresión relativa de ARNm de CTLA-4 disminuyó en pacientes con LN (p=0,044); sin embargo, la expresión relativa de ARNm de IL-37 se sobreexpresó en pacientes con LN (p=0,001) en comparación con aquellos sin LN. Hubo una sobreexpresión significativa de ARNm relativo de IL-37 y PTPN-22 en pacientes con LES activo. Pero hubo una diferencia no significativa en la expresión de CTLA-4 con la actividad de la enfermedad. El análisis de regresión reveló que los pacientes con sobreexpresión relativa de ARNm de IL-37 tenían el doble de riesgo de desarrollar nefritis lúpica (OR: 1,906; IC 95%: 1,218-2,983; p=0,005).
ConclusionesLa expresión relativa de ARNm de IL-37 fue elevada en pacientes con LES y se asoció con compromiso renal y actividad de la enfermedad. Podría considerarse como una nueva herramienta de predicción prometedora para LN. La expresión relativa del ARNm de PTPN-22 se correlacionó con la actividad de la enfermedad solo en pacientes con LES.
Systemic lupus erythematosus (SLE) is one of the most devastating multisystem autoimmune disorders characterized by the release of auto-antibodies against various cell surfaces and nuclear auto-antigens and the formation of immune complexes that lead to organ damage.1 The etiologies of SLE are multifarious, mainly due to T cell immune-dysregulation and loss of self-tolerance with increased activation of auto-reactive T cells, which consequently, stimulate autoantibody production by auto-B cells and increase the expression of pro-inflammatory cytokines that stimulate the immune response.2 Cytokines, TNF-α, IL-17a,IL-21, IL-10, and IL-15 levels, were increased in SLE patients. IL-17, IL-21 and TNF-α are correlated with active lupus nephritis (LN), poor prognosis and are considered as a potential biomarkers for diagnosis active LN.3,4
Although the pathogenesis of SLE remains unclear, the role of genetic susceptibilities has been progressively demonstrated. Currently, more than 60 risk genes have been identified.5 Moreover, epigenetic dysregulation, including DNA methylation, histone modification, and noncoding RNA, plays a role in the pathogenesis of autoimmune diseases, especially SLE, which has shed light on a new era for autoimmunity research.6
The activation and inhibition of immune regulatory mechanisms allow a proper response to external stimulation. Cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) co-receptor and protein tyrosine phosphatase non-receptor type 22 (PTPN-22) are involved in the inhibition of the T-lymphocytes activation process.7
CTLA-4 or CD152 belonging to the CD28 family, is an inhibitory receptor expressed on CD4+ CD25+ T regulatory (Treg) lymphocytes as well as on activated CD4+ and CD8+ T lymphocytes. Its inhibitory function prevents autoimmunity. So, it has an essential role in T cell-mediated autoimmune diseases like SLE.8 The PTPN-22 gene is a protein tyrosine phosphatase non-receptor type 22 encoding cytoplasmic tyrosine phosphatase that is also known as lymphoid phosphatase (Lyp) and is shown to act as an inhibitor of T-cell receptor signaling, activation, and proliferation. The transcript level of the PTPN-22 gene was approximately two to three times higher in the SLE group compared to that of the healthy group. In addition, PTPN-22 has a significant association with lupus nephritis.9
Interleukin (IL)-37, formerly named IL-1 family member 7 (IL-1F7), is an anti-inflammatory cytokine that suppresses innate inflammatory and immune responses. Furthermore, the functional analysis showed the negative involvement of IL-37 in the occurrence and pathogenesis of autoimmune diseases. In a detailed analysis of IL-37 expression in SLE patients, there was a higher relative expression of IL-37 and increased serum level in SLE patients in comparison to healthy controls.10
Further investigation uncovered that IL-37 relative expression and serum levels were fundamentally higher in patients with active SLE, mucocutaneous, and renal involvement.11 The increase in IL-37 in active SLE patients was related to inhibition of the production of inflammatory cytokines.10 Thus, IL-37 may provide a new target for studying the pathogenesis and treatment of SLE.
Until now there has been limited data about the expression of various genes among the Egyptian population, thus we conducted this study to investigate the expression and interplay among CTLA-4, PTPN-22, and IL-37 genes as negative regulators of immune response in Egyptian SLE patients to shed light on the molecular role of these three genes in the SLE, disease activity and prognosis. Also, to assess the clinical correlation between the previously mentioned genes relative mRNA expression with lupus nephritis that could represent useful biomarkers of SLE. Moreover, the selection of these pivotal genes was based on integrated bio-informatics analysis and ranking in databases.
Patients and methodsStudy setting and study populationThis study was carried out as a case-control study to evaluate the relative mRNA expression of CTLA-4, PTPN-22 and IL-37 genes among SLE patients, with and without nephritis, and healthy controls. The patients were recruited from the inpatient and outpatient of Physical Medicine, Rheumatology and Rehabilitation department as well as the Nephrology department of Suez Canal University Hospital. Laboratory tests and molecular biology techniques were done in Medical Biochemistry and Molecular Biology department, Oncology Diagnostic Unit Lab, and the clinical pathology department at Suez Canal University. The sample size was calculated to meet the statistical requirement with a 95% confidence level according to the mean prevalence of SLE worldwide and also the proportion of LN in SLE disease.
The study population was classified into two groups; fifty-eight SLE patients, diagnosed according to the American College of Rheumatology (ACR) revised updated criteria for the classification of SLE12 versus 50 age and gender matched healthy controls. In both groups, the age of the participants was more than 16 years old. The patients with other autoimmune diseases as (Rheumatoid arthritis, Spondyloarthropathies, Dermatomyositis, polymyositis, Sarcoidosis and Systemic sclerosis) were excluded.
The participants underwent careful medical history and examination. The SLE group was further classified into two groups: twenty-nine patients with lupus nephritis and 29 SLE patients without nephritis. Lupus nephritis was defined when the SLE patient had proteinuria more than 0.5g per day. Disease activity was assessed using the SLE Disease Activity Index (SLEDAI).13
All participants enrolled in the study signed informed consent, according to the regulations of the research ethics committee of the Faculty of Medicine, Suez Canal University, approval letter ID: 4311.
Laboratory assessmentLaboratory assessment was done in the form of complete blood count (CBC) (Sysmex XT 5 parts differential cell counter, Germany),serum creatinine, 24-hrs protein, c-reactive protein (CRP), complement (C3&C4) levels by (COBAS e411 Roche Diagnostics, Germany), and erythrocyte sedimentation rate (ESR) by the Westergren method. Assessment of antinuclear antibodies (ANA) by (Bio-Rad, HEp-2 cells immunofluorescence assay)and dsDNA antibodies were measured by enzyme-linked immunosorbent assay (ELISA).
CTLA-4, PTPN-22 and IL-37 gene expressionFresh whole blood samples were collected in EDTA coated tubes from all the patients and healthy controls to extract total RNA from peripheral blood mononuclear cells (PBMC) with Trizol according to the manufacturer's instructions of the QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany, cat. No.52304). NanoDrop ND-1000 spectrophotometer (NanoDrop Tech., Inc. Wilmington, DE, USA) was used to assess the quantity and quality of RNA as well as cDNA. On the thermocycler (Robocycler Gradient 96, BIOMETRA®, LA, USA), reverse transcription was performed with the High-Capacity cDNA Reverse Transcription Kit (Invitrogen/Life Technologies, USA, cat. No. Archive). The relative mRNA expression levels of CTLA-4, IL-37 and PTPN-22 were assessed by StepOne™ Real-Time PCR System (Applied Biosystem) by using Maxima SYBR Green qPCR master mix (Thermofisher Scientific, USA, cat. No. K0251) and genes’ specific primer assays (willowfort, UK) that have been designed by us: primer nucleotide sequence is used in Real-time PCR as Forward primer Sequence for CTLA-4 was 5′TGGCTTGCCTTGGATTTCAGC3′ and Reverse primer Sequence was 5′ACACACAAAGCTGGCGATGC3′, IL-37 Forward primer Sequence was 5′GATCACAAAGTACTGGTCCTGG3′ and Reverse primer Sequence was 5′TCCTTTATCCTTGTCACAGTAG3′, PTPN22Forward primer Sequence was 5′CTGTACTAGCAACTGCTCCA3′, Reverse primer Sequence was 5’TCCAGCTTCCTCAACCACAA3’ and GAPDH Forward primer Sequence was 5′CTCCTCACAGTTGCCATGTA3′, and Reverse primer Sequence 5′GTTGAGCACAGGGTACTTTATTG3 with thermal cycling protocol: initial denaturation for 10min at 95°C then forty cycles of denaturation at 95°C for15s, Annealing at 56°C for 30s and extension at 72°C for 30s followed by a melting curve analysis to exclude primer dimer. GAPDH was used as an internal reference. The results were expressed as 2−ΔΔCT.14 The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were followed for all PCR reactions.15
Statistical analysisThe data was analyzed using SPSS.25 software (SPSS Inc., Chicago, IL, USA). The data was presented as frequency and percentages for qualitative variables and as mean and standard deviation for quantitative variables. Chi-square test was used to compare categorical variables and t-test was used for quantitative variables. Spearman correlation coefficient test was used to evaluate the correlation of the relative mRNA expression of CTLA-4, PTPN22 and IL-37 with different variables. Regression analysis and Odds ratio were done to detect the ability of relative CTLA-4, PTPN-22, and IL-37 mRNA expressions to predict LN in SLE disease.
ResultsPatients’ characteristicsDemographic, clinical and laboratory data of SLE patients are shown in Table 1. The mean age of SLE patients was 31.67±9.27 years, 94.8% were females and the mean of the disease duration was 7.12±5.15 years. Other clinical parameters including disease activity and treatment taken were also listed. The laboratory characteristics of SLE patients revealed that almost all of them (91.4%) were ANA positive, and roughly 60% were anti-dsDNA positive, with elevated ESR (52.7731.44mm/h) and low hemoglobin level (10.71.2g/dl). TLC, Platelets, creatinine, C3 and C4 were also listed. Demographic, clinical, and laboratory data of the SLE without nephritis versus SLE with nephritis are demonstrated in Table 2.
Demographic, clinical, and laboratory data of the SLE patients.
| Variables | SLE patients(n=58) |
|---|---|
| Age, years, mean±SD | 31.67±9.27 |
| Females, n (%) | 55 (94.8%) |
| Disease duration, years, mean±SD | 7.12±5.15 |
| SLEDAI, mean±SD | 8.9±8.5 |
| Disease activity | |
| No activity | 13 (22.4%) |
| Mild activity | 12 (20.7%) |
| Moderate activity | 14 (24.1%) |
| High activity | 13 (22.4%) |
| Very high activity | 6 (10.3%) |
| Main system affected | |
| Renal affection | 29 (50%) |
| Musculoskeletal | 9 (15.5%) |
| Neuropsychiatric | 8 (13.8%) |
| Acute cutaneous | 8 (13.8%) |
| Cardiovascular | 4 (6.9%) |
| Laboratory findings | |
| Positive ANA, n (%) | 53 (91.4%) |
| Positive anti-dsDNA, n (%) | 38 (65.5%) |
| Low C3, n (%) | 15 (25.9%) |
| Low C4, n (%) | 12 (20.7%) |
| ESR, mm/h, mean±SD | 52.77±31.44 |
| Hb, gm/dl, mean±SD | 10.7±1.2 |
| TLC, ×103/mm3, mean±SD | 6.8±2.2 |
| Platelets, ×103/mm3, mean±SD | 168±0.1 |
| Serum creatinine, mg/dl, mean±SD | 1.1±1.06 |
| 24h proteins, g/day, mean±SD | 0.78±0.63 |
| Medications | |
| Steroids, n (%) | 41 (70.7%) |
| HCQ, n (%) | 31 (53.4%) |
| MMF, n (%) | 11 (19.1%) |
| Cyclophosphamide, n (%) | 4 (6.9%) |
| Cyclosporin, n (%) | 1 (1.7%) |
| Azathioprine, n (%) | 26 (44.8%) |
ANA: antinuclear antibody; anti dsDNA: anti double stranded DNA; C3: complement protein 3; C4: complement protein 4; ESR: erythrocyte sedimentation rate; Hb: hemoglobin; TLC: total Leukocyte count; SLEDAI, systemic lupus erythematosus disease activity index; HCQ; hydroxychloroquine; MMF; mycophenolate mofetil.
Demographic, clinical, and laboratory data of the SLE patients without lupus nephritis versus SLE patients with nephritis.
| Variables | SLE patients without nephritis(n=29) | SLE patients nephritis(n=29) | p-Value |
|---|---|---|---|
| Age, years, mean±SD | 31.89±9.05 | 31.44±9.64 | 0.856 |
| Females, n (%) | 27 (93.1%) | 28 (96.6%) | 0.553 |
| Disease duration, years, mean±SD | 7.71±5.26 | 6.51±5.06 | 0.395 |
| SLEDAI, mean±SD | 9.86±8.53 | 8.10±8.05 | 0.447 |
| Disease activity: n (%) | |||
| No activity | 4 (13.8%) | 9 (31%) | 0.092 |
| Mild activity | 9 (31%) | 3 (10.3%) | |
| Moderate activity | 4 (13.8%) | 9 (31%) | |
| High activity | 9 (31%) | 5 (17.2%) | |
| Very high activity | 3 (10.3%) | 3 (10.3%) | |
| Laboratory findings | |||
| Positive ANA, n (%) | 25 (86.2%) | 28 (96.6%) | 0.269 |
| Positive anti-dsDNA, n (%) | 17 (58.6%) | 21 (72.4%) | 0.426 |
| Low C3, n (%) | 8 (27.6%) | 7 (24.1%) | 0.764 |
| Low C4, n (%) | 5 (17.2%) | 7 (24.1%) | 0.517 |
| ESR, mm/h, mean±SD | 53.82±31.75 | 51.72±31.65 | 0.801 |
| Hb, gm/dl, mean±SD | 10.88±1.43 | 10.52±1.23 | 0.307 |
| TLC, ×103/mm3, mean±SD | 6.77±2.4 | 6.83±2.1 | 0.917 |
| Platelets, ×103/mm3, mean±SD | 161.4±116.5 | 174.9±87.5 | 0.625 |
| Serum creatinine, mg/dl, mean±SD | 0.98±0.80 | 1.24±1.03 | 0.398 |
| 24h proteins, g/day, mean±SD | 0.12±0.05 | 1.66±1.37 | <0.001* |
| Medications: n (%) | |||
| Steroids | 29 (100%) | 29 (100%) | 1.00 |
| HCQ | 17 (58.6%) | 14 (48.3%) | 0.430 |
| MMF | 5 (17.2%) | 6 (20.7%) | 0.738 |
| Cyclophosphamide | 1 (3.4%) | 3 (10.3%) | 0.300 |
| Cyclosporin | 0 (0%) | 1 (3.4%) | 0.313 |
| Azathioprine | 12 (41.4%) | 14 (48.3%) | 0.597 |
SLEDAI: systemic lupus erythematosus disease activity index; ANA: antinuclear antibody; anti dsDNA: anti-double stranded DNA; C3: complement protein 3; C4: complement protein 4; ESR: erythrocyte sedimentation rate; Hb: hemoglobin; TLC: total leukocyte count; HCQ; hydroxychloroquine; MMF; mycophenolate mofetil.
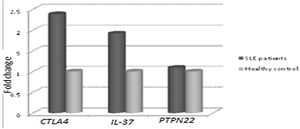
Concerning, the relative mRNA expression levels of CTLA4, PTPN22, IL-37 genes in SLE patients versus the healthy controls, as shown in Fig. 1. It was found that there was a significantly higher mRNA expression of CTLA-4 and IL-37 in SLE patients than in the healthy control group. The mean fold change of CTLA-4 gene expression was 2.380±2.138 (p=0.005) and the mean fold change of IL-37 gene expression was 1.915±1.687 (p=0.018). While there was a non-significant difference in PTPN22 gene expression in SLE patients versus the healthy control group as the mean fold change of PTPN22 gene expression was 1.088±0.817 (p=0.631).
CTLA-4, IL-37 and PTPN22 gene expression in SLE patients versus controls. There were significantly higher expression of CTLA-4 and IL-37 in SLE patients compared to control (p=0.005, and p=0.018, respectively). On the other hand, there was a non-significant difference in PTPN22 expression in SLE patients versus controls (p=0.631).
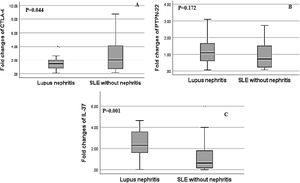
Regarding, the relative mRNA expression of CTLA4, PTPN22, IL-37 genes in SLE patients with nephritis versus SLE patients without nephritis, the relative expression of IL-37 showed a significant increase in SLE patients with LN and the fold change was greater up to 3 fold up-regulated in LN patients (2.60±1.63) versus SLE patients without nephritis (1.22±1.06, p=0.001). There was a significant decrease in the relative mRNA expression of CTLA-4 in LN patients in comparison to SLE patients without nephritis and the fold change was (1.81±1.69) in LN patients versus (2.94±2.40) in SLE patients without nephritis (p=0.044). However, there was a non-significant difference in the relative mRNA expression of PTPN-22 between both groups as the fold change was (1.23±0.81) in LN patients versus (0.94±0.70) in SLE patients without nephritis, (p=0.172) (Fig. 2).
Association of relative CTLA-4, IL-37 and PTPN22 mRNA gene expression in SLE patients with lupus nephritis (LN) versus without lupus nephritis (LN). (A) There was a significant decrease in the relative expression of CTLA-4 in SLE patients with LN in comparison to SLE patients without LN (p=0.044). (B) There was a non-significant difference in PTPN22 expression in SLE patients with and without LN (p=0.172). (C) There was significantly higher expression of IL-37 in SLE patients with lupus nephritis compared to those without lupus nephritis (p=0.001).
We evaluated the correlation of the relative mRNA expression of CTLA-4, PTPN-22 and IL-37 genes with some clinical and laboratory parameters in SLE patients, as presented in Table 3. The correlation analysis of genes expression with disease activity was performed. There was a significant positive correlation of PTPN22 and IL-37 with SLE disease activity (r=0.272, p=0.039; r=0.324, p=0.013 respectively). In addition, there was a significant negative correlation of IL-37 with low C3 level (r=0.260, p=0.049). But CTLA-4 expression was not correlated with any of the SLE studied parameters.
correlation analysis between CTLA4, PTPN22, & IL-37 expression with some clinical and laboratory parameters in SLE patients.
| CTLA4 gene expression | PTPN22 gene expression | IL-37 gene expression | ||||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| Disease duration | −0.228 | 0.094 | 0.185 | 0.175 | −0.188 | 0.170 |
| Active SLE | 0.121 | 0.367 | 0.272 | 0.039* | 0.324 | 0.013* |
| Low C3 | 0.157 | 0.239 | 0.152 | 0.255 | −0.260 | 0.049* |
| Low C4 | 0.212 | 0.111 | 0.135 | 0.313 | −0.239 | 0.071 |
| Positive anti-dsDNA | 0.047 | 0.726 | 0.036 | 0.786 | 0.101 | 0.451 |
C3: complement protein 3; C4: complement protein 4; Anti-dsDNA: anti-double stranded DNA.
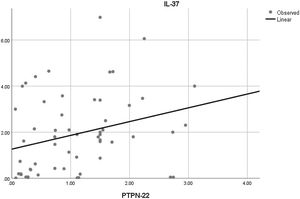
Moreover, the correlation analysis of the relative mRNA expression among the studied genes with each other was also done. There was only a significant positive correlation between the relative mRNA expression of PTPN22 and IL-37 in SLE patients (r=0.289 and p=0.028) (Fig. 3).
Regression analysis to detect the ability of relative CTLA4, PTPN22, and IL-37 mRNA expressions to predict the development of LNOver-expression of IL-37 was associated with an increased likelihood of exhibiting lupus nephritis. Patients with relative IL-37 mRNA over-expression have twice likelihood to exhibit lupus nephritis (OR=1.906, 95% CI=1.218–2.983, p=0.005). On the other hand, CTLA-4 and PTPN-22 were not associated with increased likelihood of developing lupus nephritis (OR=0.755, the 95% CI=0.567–1.006, p=0.055; OR=1.084, the 95% CI=0.510–2.301, p=0.834 respectively), as presented in Table 4.
Regression analysis predicting the effects of CTLA4, PTPN22, and IL-37 genes’ expression on the likelihood of developing lupus nephritis in SLE patients.
| r | OR (95%CI) | p-Value | |
|---|---|---|---|
| CTLA4 expression | −0.281 | 0.755 (0.567–1.006) | 0.055 |
| PTPN22 expression | 0.080 | 1.084 (0.510–2.301) | 0.834 |
| IL-37 expression | 0.645 | 1.906 (1.218–2.983) | 0.005* |
OR: odds ratio; CI: confidence interval.
SLE is considered a systemic autoimmune disease associated with the excess production of autoantibodies and immune complexes causing, injury to many organs due to dysregulation of helper T cells (Th) and regulatory T cells (Treg).16 Kidneys consider as a common organ affected by lupus, about 40% of lupus patients have lupus nephritis (LN) and about 10% of LN patients have a risk to develop end-stage kidney disease.17 So, we studied the relative mRNA expression of CTLA-4, PTPN-22 and IL-37 genes in Egyptian SLE patients and correlated their expression to LN occurrence.
Regarding genes expression among the studied SLE patientsRelative CTLA-4 mRNA expression was significantly elevated in our SLE studied patients. The level of serum soluble CTLA-4 (sCTLA-4) was found to be increased in SLE patients,18 however, there were no significant correlations between CTLA-4 gene expression and any laboratory markers or disease activity in our SLE study patients. According to that CTLA-4 may not affect exacerbations of SLE activity. Furthermore, PTPN-22 gene expression in the SLE patients showed a non-significant difference from control, but it had a significant positive correlation with disease activity. In contrast to our study results, it was reported that the level of PTPN-22 was higher in the SLE group in comparison to healthy controls.9 However, Machado-Contreras et al. found that PTPN-22 mRNA expression was reduced or absent in SLE patients who had a severe flare, and they concluded that there was no link between PTPN22 and the development of SLE in a Mexican population.19
Concerning relative IL-37 gene expression in this study, it was significantly higher in SLE patients than in control groups. Moreover, it had a significant positive correlation with low C3. Wu et al. showed that the level of C3 was positively associated with plasma IL-37 levels in SLE patients.20
Additionally, IL-37 gene expression in our SLE patients had a significant positive correlation with SLE activity. Many studies confirmed that IL-37 mRNAs expressions and serum protein levels were higher in patients with SLE than in controls and that IL-37 mRNAs and protein levels were significantly higher in patients with active diseases.10,11 Ye et al. suggested that IL-37 was associated with SLE disease activity because its anti-inflammatory effects were enhanced in the inflammatory phase and active state only. Nold et al. reported, IL-37 inhibited P38MARK phosphorylation which resulted in decreasing serum levels of pro-inflammatory cytokines and tissue damage in SLE disease.21
Regarding genes expression in SLE patients with LN compared to SLE patients without LNCTLA-4 gene expression was significantly lower among SLE patients with LN than in those without LN, the lower relative expression of CTLA-4 gene may be associated with inflammation in the kidney causing nephritis. In addition, a study conducted by Grywalska et al., confirmed that the low CTLA-4 expression and low serum level of sCTLA-4 may lead to continuous activation of T-cells causing glomerular inflammation and injury. Also, the anti-CTLA-4 monoclonal antibody (ipilimumab) that was used by oncology patients, was associated with the development of various glomerular pathologies.22
Huang et al. revealed that CTLA-4-Immunoglobulin combined with dendritic cells could have potential therapeutic effect to enhance immune tolerance and promote life quality of patients with SLE.23
Whereas there was a non-significant difference in PTPN-22 gene expression in SLE patients with LN compared to those without LN. A study by Orozco et al. discovered no significant differences in the distribution of PTPN-22 1858CT genotypes between SLE patients with and without LN.24 Another study, however, has shown that the PTPN-22 1858 C/T SNP polymorphism is associated with an increased risk of nephritis in SLE patients.25 Tizaoui et al. demonstrated that PTPN-22 have an important role in autoimmune diseases and give insights into these disease mechanisms and could be an attractive therapeutic target of several autoimmune diseases. According to the role of PTPN-22 in stimulation of T and B cells, antagonizing the effect of PTPN22 could be a novel chance to develop a proper treatment for patients with autoimmune disease as auto-reactive B and T cells inhibition is essential.26
Alternatively, in this study, there was a significantly higher IL-37 expression in LN patients compared to those without LN, also regression analysis proved that the higher IL-37 expression is considered as a significant predictor marker for the development of LN. Tawfik et al. confirmed the same results, that the serum level of IL-37 in SLE patients was higher among lupus nephritis patients.11 Moreover, Yang Y et al. detected that Small interfering RNA smashed of IL-37 in PBMCs of human renal tubular epithelial cells inhibits the kidney expression of inflammatory cytokines IL-6, TNF-α and IL-1β.27
Interestingly, a study by Ding L et al., proved that IL-37 has a protective effect in the kidney among lupus nephritis mice by reducing the deposition of the autoimmune complex as IgM, IgG, and C3, so lowering the production of inflammatory cytokines like IL-17 and IL-6 and increasing the anti-inflammatory cytokines like IL-10 and TGFβ-3, also increases the frequency of Treg cells and reduces the frequency of Th17 cells.28 Regarding other cytokines expression in SLE patients, it was interesting to show that IL-36α is significantly more expressed in Egyptian SLE patients compared to healthy controls and correlated with SLE disease activity and arthritis.29
The potentiated therapeutic effect of IL-37 is the inhibitory influence on the inflammatory microenvironment. Therefore, Genetic modification to overexpress IL-37 may enhance the therapeutic effects.30
It should be noted, PTPN-22 gene expression showed a significant positive correlation with IL-37 gene expression and up to our best knowledge, we considered ourself the first study to shed light on this correlation. Both genes have the same protective and suppressive role against the over-activity of the immune system. More studies are needed to confirm this correlation and other linked genes that communicate with both genes.
To the best of our knowledge, this study is the first to investigate the expression and interplay among CTLA-4, PTPN-22, and IL-37 genes in Egyptian SLE patients. Furthermore, the key strengths of the study included the in-depth analysis of CTLA-4, PTPN-22, and IL-37 genes expression with disease activity and renal involvement and shed light on the molecular role of these three genes in the pathogenesis and prognosis and also could represent useful biomarkers of SLE. However, it would be interesting to investigate these genes in further prospective cohort studies with large sample sizes to confirm our results in Egyptian SLE patients and establish the role of CTLA-4, PTPN-22, and IL-37 genes in the pathogenesis of SLE disease.
ConclusionRelative IL-37 mRNA expression was elevated in SLE patients and associated with renal involvement and disease activity. Relative CTLA-4 mRNA expression increased in SLE patients however decreased in LN and relative PTPN-22 mRNA expression was correlated with disease activity only in SLE patients. Finally, IL-37 can be considered as a new and promising predictive tool for lupus nephritis and disease activity in SLE patients.
Authors’ contributionsAll authors have been contributed equally to this work. They shared all in the conception and design of the study, the analysis and interpretation of data, drafting and revision of the article critically for important intellectual content. They reviewed and revised the manuscript and approved all, the final version to be published. All authors were involved in the decision to submit the manuscript for publication and had the right to accept or reject comments or suggestions.
Availability of data and materialData are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplementary information. Data will be made available on reasonable request to the author (Marwa M. Hosny).
Ethical approval informationEthical approval for the study was secured by the research ethics committee of the faculty of medicine, Suez Canal University, ID no. 4311. The study was conducted according to Declaration of Helsinki.
Consent to participateAll participants provided an informed written consent.
Consent for publicationNot required from the study participants.
FundingThere is no funding of this work.
Conflict of interestAll authors have not any competing interests.
We would like to express our great thanks to the Oncology Diagnostic Unit lab in our institution for providing the facilities for molecular biology techniques for our study. The authors would like to thank all controls and patients who participated in the study.