This study is designed to evaluate the potential influences of Mediterranean fever gene (MEFV) gene polymorphism on systemic lupus erythematosus (SLE) in a cohort of juvenile patients. A case–control study was performed on Iranian patients with a mixed ethnicity population.
Patients and methodsGenotypes of 50 juvenile cases, and 85 healthy controls were investigated for identifying M694V and R202Q polymorphism. Genotyping was done utilizing amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) to detect M694V and R202Q mutations, respectively.
Main findingsOur study indicates significant differences in the alleles and genotypes frequencies of MEFV polymorphism between SLE patients and healthy controls (P<0.05). Also, an association was found between renal involvement (50% vs. 8.3%, P=0.000, OR=0.91, 95% CI=0.30–0.278) in juvenile SLE patients and M694V polymorphism incident; But there was no association with other clinical manifestations.
Principal conclusionWe found a significant association between R202Q and M694V polymorphism of the MEFV gene and susceptibility to SLE in the studied population; However, further studies on detailed characterization of these polymorphisms’ impacts on the key elements responsible for SLE pathogenesis is of great importance.
Este estudio está diseñado para evaluar las posibles influencias del polimorfismo del gen de la fiebre mediterránea (MEFV) en el lupus eritematoso sistémico (LES) en una cohorte de pacientes jóvenes. Se realizó un estudio de casos y controles en pacientes iraníes con una población de origen étnico mixto.
Pacientes y métodosSe investigaron los genotipos de 50 casos juveniles y 85 controles sanos para identificar el polimorfismo M694V y R202Q. El genotipado se realizó utilizando amplificación refractaria sistema de mutación-reacción en cadena de la polimerasa (ARMS-PCR) y reacción en cadena de la polimerasa-polimorfismo de longitud de fragmentos de restricción (PCR-RFLP) para detectar mutaciones M694V y R202Q, respectivamente.
Hallazgos principalesNuestro estudio indica diferencias significativas en las frecuencias de alelos y genotipos del polimorfismo MEFV entre pacientes con LES y controles sanos (p<0,05). Además, se encontró asociación entre compromiso renal (50% vs. 8.3%, p=0,000, OR=0.91, IC 95%=0,30–0,278) en pacientes con LES juvenil e incidente de polimorfismo M694V; pero no hubo asociación con otras manifestaciones clínicas.
Conclusión principalEncontramos una asociación significativa entre el polimorfismo R202Q y M694V del gen MEFV y la susceptibilidad a LES en la población estudiada; sin embargo, es de gran importancia realizar más estudios sobre la caracterización detallada de los impactos de estos polimorfismos en los elementos clave responsables de la patogénesis del LES.
SLE is a chronic autoimmune disorder with multi-system organ involvement, characterized by the production of autoantibody that leads to tissue injury. It primarily affects women of childbearing age (F:M ratio 9:1), and the number of diagnosed cases is escalating rapidly.1
Moreover, comorbidities including Familial Mediterranean fever (FMF) have shown to be contributing to development of SLE.
Despite the growth of insight into the notability of environmental and hereditary factors, the etiology of SLE is yet to be understood.2 Genetics is a strong indicator of one's susceptibility to developing SLE; accordingly, the knowledge over the genes’ role and protein interactions responsible for disease development can further our understanding of the SLE pathogenesis. Investigating the association of single nucleotide polymorphism (SNP) with disease formation would identify different loci accountable for disease susceptibility.3,4 Previous case–control association studies have revealed various collaborative gene loci, including many variants in the major histocompatibility complex (MHC) coding regions, Fc- receptors, and other influential protein-coding genes.5
FMF is an autoinflammatory disorder characterized by recurrent fever attacks and abdominal, chest, and joint pains.6 Eventually, these symptoms may be followed by the most severe affliction of FMF, renal involvement, due to amyloid deposition.7 The likely explanation for the occurrence of above-mentioned comorbidity is the highly shared pathways leading to FMF and SLE.
The variants of MEFV have shown to be the causative factors of FMF and SLE. Firstly recognized in 1997, it is comprised of ten exons and positioned on chromosome 16p13.3.8 The MEFV gene encodes a protein called pyrin, also known as marenostrin, predominantly expressed in granulocytes that play a crucial function in the inflammatory response.9
So far, More than 100 MEFV gene variants (polymorphism/mutations) have been reported, most of which are in exon ten and exon two. A polymorphism in exon ten, i.e., M694V, is the most common observed variant in the Mediterranean populations and was also detected in some cases in this study.10,11 Moreover, controversy is aroused regarding the nature of the R202Q (c.605G>A) variant, observed in exon 2. It is not well-defined whether this variant is a mutation contributed to FMF, or it is only a polymorphism present in healthy individuals to.12
The aim of this study was to evaluate the clinical significance and frequency of the R202Q and M694V polymorphism in Iranian juvenile SLE cases.
Patients and methodsStudy subjectsOverall, 50 juvenile SLE cases and 85 juvenile healthy individuals were enrolled in the current study. The mean age was 12.51±3.24 years, and the mean duration of time since the onset of SLE was 2.9±3.07 years for cases. The racial distribution was 42% Fars, 30% Turks, 5% Kurd, and 9% Lor. Amongst SLE cases, 42 were females, and 8 were males (F:M ratio of 5.25:1). The majority of lupus patients, regardless of MEFV polymorphism status, were females. In order to minimize the effects of unknown population stratification, the age, sex, and ethnic-matched healthy control group was included in this study.
All patients fulfilled at least four American College of Rheumatology criteria for SLE.13 The Healthy subjects did not have any clinical indications or history of autoimmune and genetic diseases. The laboratory and clinical records of all individuals were recorded using a standard form.14
Informed consent was attained from the parents prior to their enrolment in the study regarding the identified data to be published and also, the children assented. All procedures were conducted after informed consent was obtained from patients and in accordance with the local ethics committee.
DNA extraction and genotypingDNA extraction was done from the whole blood of each patient, as explained previously.15 The variations of MEFV M694V (c.2080A>G, rs61752717) and R202Q (c.605G>A, rs224222) SNPs were determined by ARMS-PCR and PCR–RFLP, respectively.
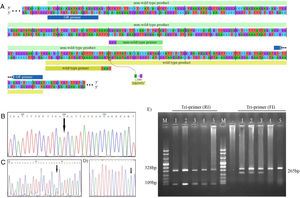
M694V SNP identification using ARMS-PCRThe primers for ARMS-PCR were designed using Primer1 online software. The criteria suggested by Medrano et al. were considered for designing the primers. The Primer-Blast tool was employed for assurance of specificity. The allele-specific primers contained a mismatch at the −3 position of the 3′ terminus (Fig. 1A). The sequence of primers used for genotyping and the amplicon size are reported in Table 1.
Genotyping of M694V polymorphism. (A) The schematic illustration of M694V polymorphism site on MEFV gene and the primers designed for genotyping. The chromatograms representing normal homozygote (B), heterozygote (C), and M694V homozygote (D) states which were perfomed for the validation of genotyping. (E) Genotyping of M694V polymorphism by ARMS-PCR. Lane M, molecular-weight size marker; Lane 1, 2, and 3 indicate AG genotype and lane 4 and 5 indicate AA genotype.
ARMS-PCR primers used for the identification of M694V polymorphisms.
| Polymorphism | Primer | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| M694V | IF | GCTACTGGGTGGTGATAAGGG | 265 |
| IR | GACGCCTGGTACTCATTTTCCTTAAT | 109 | |
| OF | TCCTGGGAGCCTGCAAGAC | 328 | |
| OR | ACTGGACAGATAGTCAGAGG | 328 |
IF, inner forward; IR, inner reverse; OF, outer forward; OR, outer reverse.
Wild-type and non-wild-type products were amplified in two separated PCR reactions; outer forward and outer reverse primers were present at both reactions. Each allele-specific primer was added to a single reaction.
The amplification conditions for the tri-primer reaction containing the non-wildtype primer were as follows: initial denaturation at 94°C for 2min, 35 cycles at 94°C for 30s, 59°C for 30s, and 72°C for 40s, the reaction was ended after a final extension at 72°C for 10min. The PCR conditions for the other tri-primer reaction were the same, except the annealing temperature set at 61°C.
The amplified PCR products were mixed with loading buffer and further visualized by electrophoresis in 2 percent agarose gel stained with ethidium bromide under UV light. ARMS-PCR band patterns for cases 1–5 are shown in Fig. 1E.
Validation of ARMS-PCR assayPCR products were purified, and automated DNA sequencing was carried out for ten percent of samples randomly. Representative of each genotype are present as chromatograms in Fig. 1B–D.
Genotyping of R202Q polymorphism by RFLPR202Q polymorphism of the MEFV gene were analyzed using RFLP-PCR. Briefly, forward and reverse primers flanking the polymorphism site were employed to amplify a 501bp fragment of the MEFV gene. Description of primers and products are addressed in Table 2. The PCR conditions were 1min at 94°C, 35 cycles of 30s at 94°C, 36s at 58°C and 50s at 72°C, with a final extension at 72°C for 3min. The PCR product was further digested with 6U PvuII (Thermo Scientific™ Fermentas) at 37°C in a water bath for 16h. In the case of allele G present at the polymorphism site, no cleavage occurs. If allele A is present, the restriction enzyme cleaves the product into 305 and 196bp fragments. The samples’ genotype was determined by observing bands of different lengths in 2% agarose gel stained with the green viewer under UV light.16 The GG, GA, and AA genotypes result in one, three, and two bands on agarose gel, respectively.
Statistical analysisThe statistical departure of the two SNPs from the Hardy-Weinberg equilibrium (HWE) was computed. The tests were performed independently for cases and especially for the controls. The genotype test of 3×2 contingency tables and the comparison of the minor allele frequencies (MAF) were performed utilizing χ2 and Fisher's exact test, respectively. The tests of deviation from HWE and both genotypic and allelic associations were carried out using plink software. Odds ratio (OR) and 0.95 confidence interval (CI) were measured for both SNPs. The empirical significance levels were generated using the basic (adaptive) permutation procedure for multiple testing correction to account for the relatively small sample size. Maximum permutations number of 1,000,000 times was set for both of the SNPs.
To check the null hypothesis that the categorical data consisting of laboratory and clinical records are proportionately distributed between the carriers and non-carriers of the mentioned polymorphism, χ2 was performed using SPSS software. Also, population stratification was done for the SLE cases.
ResultsFrequency of M694V and R202Q in SLE patients and controlsThe genotypes frequency of R202Q and M694V polymorphism and the significance level of compliance to HWE are listed in Table 3. No deviance from HWE was detected in cases and controls, separately (P>0.05).
Distribution of MEFV genotyping and alleles frequency in juvenile subgroups of patients and control groups.
| SNP ID | Position (chr16:) | Population | Allele1 2 | Genotype | Alleles | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | HWE | P-value | Permutation (EMP1) | n (%) | P-value | OR | 95%CI | Permutation | |||||||
| 1 1 | 1 2 | 2 2 | 1 | 2 | 1 vs. 2 | ||||||||||
| R202Q (rs224222) | 3254463 | SLE | G A | 37 (74) | 11 (22) | 2 (4) | 0.28 | 2.13×10−3 | 5.1×10−4 | 85 (85) | 165 (97) | 4.55×10−4 | 5.824 | 2.047–16.57 | 1.12×10−3 |
| Control | 80 (94) | 5 (6) | 0 (0) | 1 | 15 (15) | 5 (3) | |||||||||
| M694V (rs61752717) | 3243407 | SLE | A G | 33 (66) | 14 (28) | 3 (6) | 0.37 | 4.79×10−4 | 4.71×10−3 | 80 (80) | 162 (95) | 1.23×10−4 | 5.062 | 2.137–11.99 | 7.34×10−3 |
| Control | 78 (92) | 6 (7) | 1 (1) | 0.16 | 20 (20) | 8 (5) | |||||||||
HWE, Hardy–Weinberg equilibrium.
The minor allele frequency (MAF) of R202Q polymorphism showed a statistically significant difference between cohorts of cases and controls in juvenile patients (15% vs. 2.9%, P=0.0004, OR=5.824, 95% CI=2.047–16.57). Moreover, a significant association was detected for the M694V polymorphism (20% vs. 4.1%, P=0.0001, OR=5.062, 95% CI=2.137–11.99).
Regarding the R202Q polymorphism, the ratio of cases with genotypes of GG, AG (heterozygote), and AA (homozygote) were 74%, 22%, and 4%, respectively. Considering the M694V variant, that of AA, AG (heterozygote), and GG (homozygote) were 66%, 28%, and 6%, respectively.
Clinical characteristicsThree of the juvenile patients were compound heterozygous for M694V and R202Q polymorphism. Subphenotypes were mainly renal involvement, positive ANA and anti-dsDNA, CRP titer elevation, and secondary antiphospholipid antibody syndrome (APAS).
The clinical presentation among SLE cases was analyzed. Amongst which renal involvement has proven to be more prevalent (significantly different) in carriers of M694V, compared to non-carriers (50% vs. 8.3%, P=0.000, OR=0.091, 95% CI=0.030–0.278). No significant association was found between other clinical manifestations of SLE patients and M694V variant.
Age and population stratificationsThe distribution of M694V alleles was significantly different concerning the age of disease onset (8.16±3.87 vs. 10.44±2.61 years, P=0.012). However, that was not the case for the R202Q carriers vs. non-carriers (6.89±3.15 vs.10.64±2.73 years, P=0.191).
The statistical analysis of individuals classified by ethnic backgrounds showed a significant difference for allelic presence in all the groups (P<0.05).
Discussion and conclusionsHeterozygosity of MEFV variations has shown to affect the development of inflammatory disorders, including rheumatoid arthritis, Behcet's disease, Crohn's disease and multiple sclerosis, in particular, the M694V variant as one of the most prevalent polymorphisms in various rheumatoid diseases.17 To the best of our knowledge, no single study with thorough association analysis has been established for the M694V and R202Q polymorphisms in Iranian SLE cases;18–20 therefore, we decided to focus on these polymorphism to fill the gaps.
The significant findings revealed in this study were as follows:
The frequencies of MEFV polymorphisms showed a statistically significant difference between the cases and the matched healthy controls in both R202Q and M694V SNPs.
Moreover, the permutation (adaptive) tests were carried out to account for the relatively small sample size. The empirical significance values confirmed the associations.
The M694V polymorphism carriage rate in the cohort of SLE patients was 32.4%. The M694V polymorphism carriage is likely a risk factor for renal involvement in juvenile SLE patients, considering each individual's clinical characteristics. The genotype distribution analysis demonstrates that M694V manifests itself in homozygote and heterozygote states. No significant differences were observed for other clinical signs in the cohorts. Our result contrasts the possible modifying role of MEFV polymorphisms on the clinical expression, as suggested by Y. Shinar et al.17
On the other hand, the underlying mechanism that explains the development of renal involvement in the case of M694V polymorphism presence, previously ascribed by Deniz et al.21 is in agreement with our results.
Furthermore, it has also been suggested that M694V polymorphism carriage may worsen clinical symptoms in individuals with low-grade inflammation.22 Detailed examination of specific manifestations, such as peritonitis, was not carried out on our patients; Hence, future studies focusing on the presence of peritonitis along with data on the incidence of serosal tissue invasion in SLE patients may provide more reliable and conclusive results.
R202Q (605G>A) has been reported as a common polymorphism and has also been associated with FMF causative rare mutations in Mediterranean populations.23 There is controversy on the essence of R202Q SNP; Besides other polymorphism, it is widely debated that whether SLE is a possible ramification of R202Q. Some studies define the nature of this SNP as a disease-causing mutation, while others, including a study conducted by Aldea et al. indicate R202Q as a frequent polymorphism.24 Even though the rationale behind such a discrepancy is not evident, ethnicity might be a contributing factor. The other plausible elucidation for the inconsistency of the reported results are the low numbers of samples.
In our study, the incidence of R202Q was statistically different between the two cohorts. Here however, the R202Q allelic frequency and genotype distribution were associated with the development of SLE. Unlike the M694V, there was no association between the R202Q SNP and particular clinical manifestations in SLE cases.
In the previous studies, in which R202Q was found to be a contributing factor for the development of SLE, it has been suggested that R202Q is influential in the heterozygous state,25 with a similar pattern observed in this study. However, these findings contrast to an article published by Ozturk et al.26 Systematic screening in patients with FMF-like disease for the MEFV gene polymorphism disclosed a significant proportion of heterozygous state, which depended on patient ancestry. It was observed in low frequency in the Mediterranean countries (20–25%) rather than the considerably higher frequency in other populations (80–85%).27
In homozygote R202Q cases, two of them also had M694V polymorphism introduced into the MEFV gene, one of whom was homozygote. The clinical manifestations of the homozygote patient were the antinuclear antibody (ANA) positive, renal involvement, anti-dsDNA positive, photosensitivity, malar rash, thrombocytopenia, hypocomplementaemia, and proteinuria.
The clinical manifestations of the homozygote R202Q/heterozygote M694V were renal involvement, positive anti-dsDNA, photosensitivity, malar rash, low complement and positive C-reactive protein (CRP).
These results suggest that R202Q may play a role in SLE pathogenesis along with other etiological factors and clinical features in SLE cases. However, the view of resultant downstream protein network remodeling is not entirely evident, and it may be possible to explain these phenomena by differential regulation in the transcription of genes.28
Additionally, three juvenile patients were compound heterozygote for the M694V/R202Q polymorphisms, and their clinical manifestations conformed to the criteria for SLE diagnosis.13
Population stratification was performed for the cases based on different races, and subsequently, all of them showed significant differences in allele frequencies between patients and controls (P<0.05). Therefore, it can be deduced that the ancestry was not a confounding factor in our population, and the SLE manifestations in our patients are associated with allele frequencies of MEFV polymorphisms in the patients and not because of their genetic ancestry. Permutation-based analysis was carried out to mitigate the potential bias as a result of the small sample size, and considering the limitation of accessing samples from Mediterranean ancestry patients, there may be unavoidable bias in the interpretation of the results.
Considering the preliminary findings regarding the association of the R202Q and M694V with SLE in this study, it is necessary to investigate further the role of these polymorphisms in detail via robust methods, including genome-wide association studies. In conclusion, we found that these MEFV polymorphisms could be additional genetic susceptibility factors in SLE regardless of their age of onset. Even in a heterozygote state in the juvenile, these polymorphisms can cause more severe symptoms. Also, our study shows that in juvenile patients, M694V polymorphism may lead to the early onset of the SLE disease (P=0.012).
Though this study should be considered a preliminary finding, more extensive independent studies with a broader spectrum of MEFV variations are essential to define its exact role in this disease. Analysis of these subgroups can represent the whole Iranians with mixed ethnicity. Additionally, the frequency of alleles and genotypes may vary between populations; therefore, association studies of the MEFV gene in SLE patients in different strata can lead to an accurate understanding of the MEFV polymorphism's role on SLE susceptibility and help to develop new treatments. Further research in this field would possibly aid the diagnosis of SLE as a reliable complement for differential diagnosis to distinguish it from other diseases with autoinflammatory features.
FundingThis study was funded and supported by Deputy of Research of Shahid Beheshti University.
Conflict of interestThe authors declare they have no conflict of interest.
The authors are very grateful to Mofid Children's Hospital for the excellent technical assistance and to the blood donors for their participation in this study.