Osteoarthritis (OA) is a complex disease caused by the interaction of multiple genetic and environmental factors. This review focuses on the studies that have contributed to the discovery of genetic susceptibility factors in OA. The most relevant associations discovered until now are discussed in detail: GDF-5, 7q22 locus, MCF2L, DOT1L, NCOA3 and also some important findings from the arcOGEN study. Moreover, the different approaches that can be used to minimize the specific problems of the study of OA genetics are discussed. These include the study of microsatellites, phenotype standardization and other methods such as meta-analysis of GWAS and gene-based analysis. It is expected that these new approaches contribute to finding new susceptibility genetic factors for OA.
La artrosis (OA) es una enfermedad compleja en la que diferentes factores ambientales interactúan con múltiples factores genéticos. Esta revisión se centra en los estudios que han contribuido a descubrir los factores genéticos de susceptibilidad a la OA. También se tratan con detalle los loci más relevantes en la actualidad, como GDF-5, el locus en el cromosoma 7q22, MCF2L, DOT1L, NCOA3 y los provenientes del estudio arcOGEN. Además, se discuten las diferentes aproximaciones que pueden servir para minimizar los problemas específicos del estudio de la genética de la OA. Entre ellas se encuentran la estandarización de los fenotipos, el estudio de microsatélites y también el uso de otras estrategias de estudio, como metaanálisis de GWAS y análisis basados en genes. Mediante estos nuevos enfoques se espera contribuir al descubrimiento de nuevos factores genéticos de susceptibilidad a la OA.
The first studies to show a genetic component in osteoarthritis (OA, osteoarthritis) have been twin sibling studies, relative risk among sibling studies and familial aggregation. Al of these allows an estimate of heritability, quantifying the importance of genetic factors in the disease. Heritability estimates in OA vary in function of the different studies consulted. However, it might be said that knee OA heritability is around 40%, hand OA is situated at 65% and hip OA is at 60%. Spinal OA presents the highest heritability, 70%.1 These studies have led to others, oriented to discovering OA susceptibility factors.
Linkage StudiesLinkage studies evaluate the co-segregation of genetic markers with the disease in families with multiple cases of OA. Linkage is seen when several affected members share the same allelic variant for a marker. Once the genetic region segregating with the disease has been localized, this is saturated with more markers until the linkage region is shortened and genes may be prioritized according to function. Linkage studies in OA were carried out in different cohorts with different phenotypes. Hand OA studies were carried out in Framingham (USA), Iceland and Finland. Knee/hand OA studies were carried out in the United Kingdom and the GARP Study in the Netherlands. In this form, different linkage regions in which the most likely candidate genes for a case control study of association and identify the associated polymorphisms were identified. In this way MATN3, IL4-R and the IL-12–4 cluster were identified in OA; however, most of these loci have not been posteriorly confirmed. The difficulty of progressing from linkage to the identification of susceptibility genes is due to the fact that the linkage regions are typically very large and identifying the susceptibility variant is complicated. Additionally, the loci detected have a weak effect and the size of the simple collections employed was small, making it possible for many of these results to be false positive.
Studies of Candidate GenesCandidate gene studies are based on the knowledge of the pathogenesis of OA and select genes that, due to their functional role, might be relevant for disease susceptibility. In this manner, OA candidate gene studied are those that codify for components of the extracellular matrix, anabolic or catabolic regulatory proteins of the extracellular matrix and for inflammatory mediators. Once selected, case–control gene association studies are carried out in which the allelic frequencies of the polymorphism of interest are compared, in non-related individuals with OA and disease-free controls. One of the characteristics of these studies is their capacity to detect small effects with ease. In addition, it is easier to obtain other sample collections with a relatively large size. Candidate gene studies largely detected associations, which were not later confirmed in GWAS, such as: PTGS-2, DIO-2 o FRZB.5–7 Only one of the findings of these studies, GDF-5, has been later replicated, reaching the significance level required in GWAS.8,9
Genome Wide Association StudiesGWAS allow the study of all of the genomes’ gene variability without a prior hypothesis thanks to the existence of linkage disequilibrium (LD). The organization of the genome into LD blocks allows a representation of the genomic region by selecting some informative SNPs (tagSNPs). The characteristic tagSNPs for a region are known thanks to the HapMap project, which studied a great number of SNPs in different populations. Based on this information, the high density genotype arrays provide almost complete coverage of the genome variability. The arrays used in most of the OA GWAS contain around 500 000 SNPs and, in the case of the arcOGEN study, the most powerful performed in OA, an array covering 610 000 SNPs.10,11 One of the main characteristics of GWAS is the significance threshold considered after correcting by multiple tests (P<5×10−8). This threshold is provided by the estimate that there are approximately 106 independent SNPs in the human genome (0.05/106=5×10−8) and have the advantage of reducing the rate of false positives. This significance threshold significance requirement allows the researcher to obtain solid results and, therefore, replicate them without discovering other genetic factors with a more modest effect. This degree of demand has been a great stimuli for the development of collaborative studies and meta-analysis to increase the statistical power through the increase in the size of sample collection. The power of a GWAS depends, in addition, to the effect size of the causal SNPs and the frequency of their alleles. Thanks to the GWAS and the GWAS meta-analysis, OA susceptibility genes have been progressively discovered, although the number of loci is relatively low in comparison to other complex diseases. The first solid findings were the GDF-5 gene, the 7q22 locus and MCF2L.9,12,13 More recently, the loci discovered in the arcOGEN study (GNL3, GLT8D1, ASTN2, FILIP1-SENP6, KLHDC-5-PTHLH, CHST11, TP63 and SUPT3H-CDC5L) have been incorporated, as well as other loci such as DOT1L, NCOA3 and ALDH1A2.11,14-16 It is suspected that the heterogeneity of OA phenotypes, the insufficient collection sample sizes and the design of GWAS themselves may have contributed to these insufficient results. All of these factors are considered below.
Specific Problems in Osteoarthritis Gene StudiesOne of the problems faced in the genetic study of OA is the heterogeneity in the phenotypes of OA and the way in which they are considered in different studies combining in meta-analysis or collaborative endeavors. This variability reduces the statistical power in association studies. Therefore, the standardization of patient phenotype has been recommended for future GWAS. The stratification by gender, age, BMI and presence of radiographic or symptomatic OA has been proposed. In radiographic OA (ROA), phenotypes must be defined in relation to another series of characteristics such as the number of osteophytes (ROA of the hands and knees) and the presence of joint space narrowing (hip OA).17 Another way of increasing the detection power of the new variants is to center the GWAS on the study of endophenotypes, which are phenotypes closer to the biologic cause of the disease then the disease itself with its spectrum of signs and symptoms. It is considered that endophenotypes allow the researchers to obtain more homogeneous patients and have a more direct relationship with genetic factors. In this manner, the power of the studies can be increased. The problem is the need to obtain the simple collections where they were studied. In OA, endophenotypes are characteristics of the cartilage or the form of a joint. One example of endophenotype is joint space narrowing (JSW). Its use in GWAS led to the discovery of the hip OA susceptibility gene, DOT1L.14 Another endophenotype, joint pain, has led to the discovery of PACE-4 and TRPV1 as possibly associated loci, although this has not yet reached statistical significance at the GWAS level.18,19
But not only is the selection of patients important. But also the definition of controls. OA is a late onset disease, greatly prevalent in the population, and therefore the use of population controls is widely discussed. If the cases included in the genetic studies are selected on the basis of a radiographic criterion, controls should, ideally, also be selected in this way. However, study design does not always allow it due to the difficulty often presented of having to justify the x ray examination of a healthy individual. In the first phase of the arcOGEN study, researchers evaluated how the use of population controls or OA free controls influence GWAS results. OA free controls in this study, also known as «supercontrols», were collected from the Twins UK collection of twin siblings, free of OA and selected radiographically according to a Kellgren–Lawrence<2 score. In all of the OA susceptibility loci analyzed in the arcOGEN study, only GDF-5 had a larger effect and more significant P value when using OA free controls. In the rest of the analyzed SNPs, results were identical or in some cases even better, when using population controls. Due to this, it was concluded that using OA free controls does not lead to a detectable improvement in the results of an association. However, this result cannot be considered definite because many of these associations had been discovered using population controls.20
The populations included in the studies are another source of heterogeneity. It has been proven that the same genetic factor has unequal effects on disease susceptibility in different populations. This is clearly reflected in genetic studies performed in European and Asian populations. Thus, the GWAS associations of HLA class ii/iii and DVWA are specific for the Japanese population and were not replicated in the European population. Joint specific (knee, hands, hip OA) gene susceptibility factors, as well as gender must be taken into account. This compartmentalization complicates the collection of large sample collections. It is precisely the size of the samples of OA patients available which constitutes a limiting factor. For example, the meta-analysis which identified the locus 7q22 association with knee OA included 6709 cases and 35 909 controls. However, the statistical power of this study needed to discover small effect variants (OR=1.10–1.15) was not enough. 7000 more samples would be needed to detect 1the effect of SNPs with a minor allele frequency (MAF)=20% and an OR of 1.15. Therefore, available collections are insufficient to identify modest effects.
Most OA association studies are case–control designs, although some cohorts exist. These follow a representative group of the population through time, evaluating the appearance of new cases of OA. The positive aspects of this design are its representativeness, the control over data collection according to established protocols and the absence of bias, because the demographic and environmental parameters are clearly defined and are the same as in cases as in controls. However, its main limitation is the difficulty in starting the study with a great number of individuals to follow along many years in order to gather a lot of cases.
On the other hand, GWAS have limitations due to incomplete coverage provided by the genotyping arrays. SNPs with a frequency of <5%, with a weak LD in relation to other SNPs, cannot be studied confidently. This lack of coverage is greater for rare genetic variants (MAF<0.5%), some complex copy number variants (VNTR), an microsatellites with multiple alleles and a high mutation level in the population.21 Another of the GWAS limitations, in this case related to result interpretation, is that associated SNPs do not have to be the variants that cause disease. In the past years, technology has been developed that allows for genome resequencing, or that related to the genome expressed as proteins (the exome), making it possible in this way to recover all of the information lost in GWAS with SNPs arrays and making it easier to identify polymorphisms that cause an increase in susceptibility. The latter objective has also been facilitated with the ENCODE project (ENCyclopedia Of DNA elements), which provides information on the functionality of all of the segments of the human genome.22
In summary, 2 large groups of OA associated loci may be considered:
- 1.
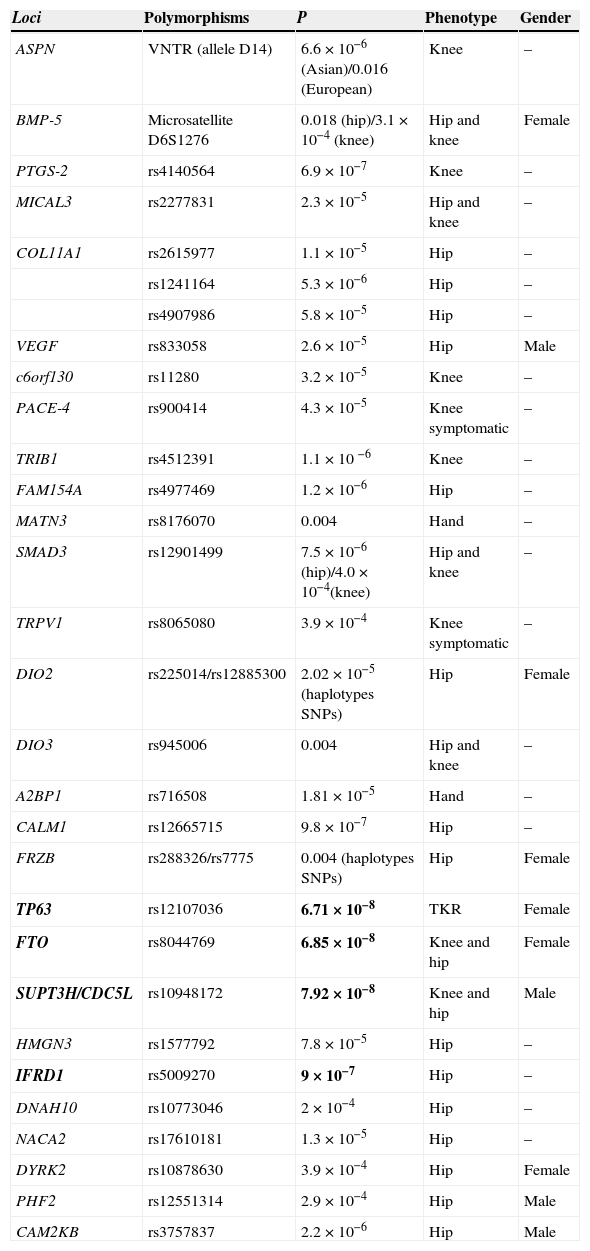
Loci possibly associated with OA but which have not reached an association level required in GWAS (Table 1).
Table 1.Loci Associated to Osteoarthritis That Does not Reach the Association Level Required in GWAS.
Loci Polymorphisms P Phenotype Gender ASPN VNTR (allele D14) 6.6×10−6 (Asian)/0.016 (European) Knee – BMP-5 Microsatellite D6S1276 0.018(hip)/3.1×10−4 (knee) Hip and knee Female PTGS-2 rs4140564 6.9×10−7 Knee – MICAL3 rs2277831 2.3×10−5 Hip and knee – COL11A1 rs2615977 1.1×10−5 Hip – rs1241164 5.3×10−6 Hip – rs4907986 5.8×10−5 Hip – VEGF rs833058 2.6×10−5 Hip Male c6orf130 rs11280 3.2×10−5 Knee – PACE-4 rs900414 4.3×10−5 Knee symptomatic – TRIB1 rs4512391 1.1×10 −6 Knee – FAM154A rs4977469 1.2×10−6 Hip – MATN3 rs8176070 0.004 Hand – SMAD3 rs12901499 7.5×10−6(hip)/4.0×10−4(knee) Hip and knee – TRPV1 rs8065080 3.9×10−4 Knee symptomatic – DIO2 rs225014/rs12885300 2.02×10−5(haplotypes SNPs) Hip Female DIO3 rs945006 0.004 Hip and knee – A2BP1 rs716508 1.81×10−5 Hand – CALM1 rs12665715 9.8×10−7 Hip – FRZB rs288326/rs7775 0.004 (haplotypes SNPs) Hip Female TP63 rs12107036 6.71×10−8 TKR Female FTO rs8044769 6.85×10−8 Knee and hip Female SUPT3H/CDC5L rs10948172 7.92×10−8 Knee and hip Male HMGN3 rs1577792 7.8×10−5 Hip – IFRD1 rs5009270 9×10−7 Hip – DNAH10 rs10773046 2×10−4 Hip – NACA2 rs17610181 1.3×10−5 Hip – DYRK2 rs10878630 3.9×10−4 Hip Female PHF2 rs12551314 2.9×10−4 Hip Male CAM2KB rs3757837 2.2×10−6 Hip Male TKR: Total knee replacement.
In bold the loci located near the threshold of association of GWAS (P<5×10−8) in Europeans. These loci come from the arcOGEN study and the recent meta-analysis of hip OA. Evangelou et al.15 makes reference to the meta-analysis of hip OA and not to the table.
- 2.
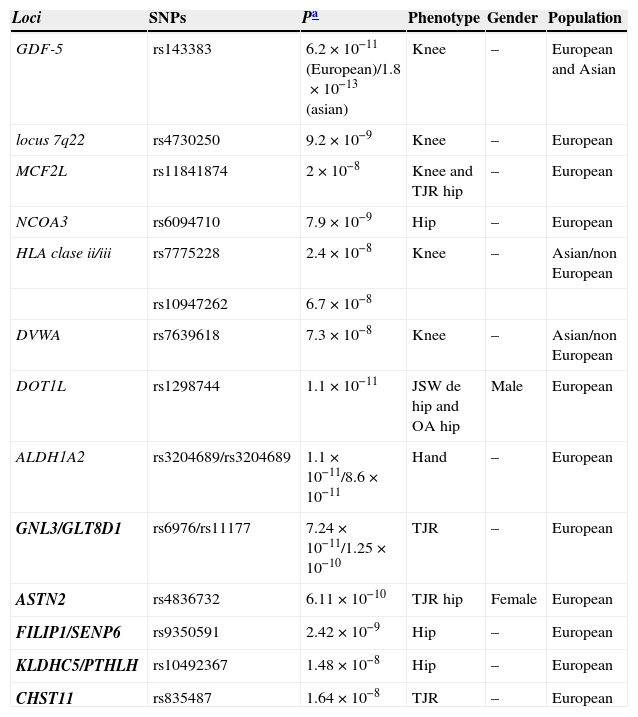
Loci which have reached a level of association with GWAS (P<5×10−8) and therefore are consistently associated with OA (Table 2).
Table 2.Loci Associated With Osteoarthritis That Reach the Association Level Required in GWAS.
Loci SNPs Pa Phenotype Gender Population GDF-5 rs143383 6.2×10−11(European)/1.8×10−13(asian) Knee – European and Asian locus 7q22 rs4730250 9.2×10−9 Knee – European MCF2L rs11841874 2×10−8 Knee and TJR hip – European NCOA3 rs6094710 7.9×10−9 Hip – European HLA clase ii/iii rs7775228 2.4×10−8 Knee – Asian/non European rs10947262 6.7×10−8 DVWA rs7639618 7.3×10−8 Knee – Asian/non European DOT1L rs1298744 1.1×10−11 JSW de hip and OA hip Male European ALDH1A2 rs3204689/rs3204689 1.1×10−11/8.6×10−11 Hand – European GNL3/GLT8D1 rs6976/rs11177 7.24×10−11/1.25×10−10 TJR – European ASTN2 rs4836732 6.11×10−10 TJR hip Female European FILIP1/SENP6 rs9350591 2.42×10−9 Hip – European KLDHC5/PTHLH rs10492367 1.48×10−8 Hip – European CHST11 rs835487 1.64×10−8 TJR – European JSW: joint space widening; TJR: total joint replacement.
In bold the loci of the arcOGEN study.
This section will deal with the most frequent OA associated loci.
GDF-5, the Only Confirmed Candidate GeneGDF-5 was the first locus associated at a GWAS level in European and Asian patients. A population study in Japan discovered the association of a SNPs (rs143383, C/T) in the 5′ UTR region of the gene. This association was observed with hip OA (P=1.8×10−13) and, with a lesser magnitude, in knee OA.8 Later studies confirmed the association of rs143383 in Asian and European persons. Additionally, a meta-analysis of European and Asian cohorts found an association of rs143383 at a GWAS level with knee OA (P=6.2×10−11).9,23 This same SNPs was associated with diverse phenotypes more or less related with OA such as hip dysplasia, bone size, female risk of vertebral fracture and height.24,25 In relation with the functional role of this gene, it was well known that GDF-5 is a member of the bone morphogenic proteins (BMP) and has a key role in skeletal development, chondrogenesis, diartrodial joint formation and bone and cartilage repair processes.26,27 The rs143383 SNPs influences GDF-5 transcriptional activity with one of its alleles, the T allele associated with OA, showing a reduced expression in cartilage. Another 2 polymorphisms of GDF-5 were later described, modifying its expression and contributing in their association to OA, being in LD with rs143383.23 The expression of GDF-5 is also modified by a series of trans regulators that interact with the rs143383 in the 5′ UTR region in addition to being a locus subjected to epigenetic regulation.28,29 Undoubtedly, GDF-5 is the genetic factor most consistently demonstrated in OA genetic studies and with a clearly established functional role in joint cartilage biology.
The 7q22 Chromosome Locus With Its 6 GenesThe second locus associated at the GWAS level in Europeans was the chromosome 7q22 locus. This is a large locus occupying more than 500kb, and containing over 400 SNPs and 6 different genes: PRKAR2B, HBP1, COG5, GPR22, DUS4L and BCAP29. The association of this locus with knee OA was discovered in a GWAS which after different study phases of 500,000 SNPs led to a single SNPs, rs3815148, with a GWAS level of association (OR=1.14, p=8×10−8).10 A meta-analysis was later performed with the pooled data from 4 GWAS done in the European population followed by another done in 9 additional European collections. In this study, rs4730250 (DUS4L) was the SNPs most associated both in the initial phase as in the global analysis of all the collections (OR=1.17, p=9.2×10−9).30 Functional locus studies centered on discovering which of the 6 genes was the best functional candidate seemed, at least initially, to suggest that it was GPR22, mainly because it was a SNPs localized upstream of the gene and largely linked with rs3815148 was associated with changes in the expression of the gene in lymphoblasts. However, its role has later been questioned because it is almost not expressed in cultured human chondrocytes or in cartilage samples. In contrast, immunohistochemical expression of GPR22 in superficial layer chondrocytes of a murine OA model, as well as the overexpression of GPR22 in vitro stimulates chondrocyte hypertrophy and accelerates calcification.31 More recent experiments have placed HBP1 as the best candidate gene to explain the association. This conclusion was reached analyzing the differential allelic expression of the 6 cartilage genes in patients with OA. This is an important gene in the Wnt signaling pathway.32 However, this is still an unresolved issue and nobody has proposed a detailed mechanism such as the one described for GDF-5.
The arcOGEN Study (Phases 1 and 2) and Its 5 LociThe arcOGEN Project is the most ambitious to date in this field and constitutes a great effort to clarify the gene basis of OA susceptibility. In its first phase, 3177 cases were studied in the United Kingdom (knee and hip OA) and 4894 controls. The identification of some associated loci was carried out in this phase, none of them at the GWAS level and none of them later confirmed. However, this phase of arcOGEN was used to analyze the genetic structure of OA and this result was interesting indeed. Its structure is that of a polygenic disease, such as most complex diseases, and not oligogenic, as postulated before the GWAS. This means that there are very likely many associated polymorphisms, each one with a small effect on predisposition to disease and that are additionally spread out homogeneously throughout the genome. In contrast with other complex diseases, in which many polymorphisms with a small effect are found close to some with marked effect, there is no evidence of great effect polymorphisms in OA.
In the second phase of arcOGEN (with a total 7410 cases of knee and hip OA, and 11,009 controls) 5 loci were identified with a GWAS significance level and another 3 came very close. Therefore, this study constitutes the greatest advancement in the identification of genetic susceptibility factors for OA. These results were obtained combining the GWAS from the arcOGEN samples with the in silico replication in a similar number of cases and controls of 6 European collections and the de novo replication, in another cohort from the United Kingdom. The 2 most associated SNPs, rs6976 and rs11177, represent a single signal associated with OA and total knee, as well as hip, replacement. One of these SNPs, rs6976, is situated in the 3′ UTR region of GLT8D1, and another, rs11177, is a non-synonymous SNPs localized in the 3 exon of GNL3, making any of these genes responsible for the association. The other 4 loci that reached GWAS significance were associated to hip OA and have been described as: ASTN2, FILIP1-SENP6, KLHDC5-PTHLH and CHST11, although there are yet not studies that determine the causality of any of these genes. The 3 loci that showed association close to the P<5×10−8 threshold have been described as: TP63, FTO and SUPT3H-CDC5L11 (Table 2). It must be pointed out that, as in the first phase of arcOGEN, no GWAS association between GDF-5 and the 7q22 locus was seen, in spite of the fact that the associations are solid. This lack of replication has been attributed to a combination of different factors: Insufficient statistical power due to the simple size in the discovery phase and the SNPs’ allelic frequencies and, in GDF-5, the absence of genotyping of rs143383. However, a tagSNPs of rs143383, rs4911494 (r2=0.94), did not show an association either. The function of some of the loci associated to joint cartilage in the second phase of arcOGEN is unknown, if any, as is the case of ASTN2. Others are a little better known, such as GNL3, which was expressed in chondrocyte precursor mesenchymal cells and in osteoarthritis chondrocytes. COL12A1, which codes for a collagen protein, is close to the FILIP1-SENP6 genes, which forms a part of the extracellular matrix. In relation to another locus, the PTHLH gene codifies a protein related to parathyroid hormone (PTHrP), which is highly expressed in the cartilage of OA patients and participates in the development of subchondral bone. CHST11, on its part, codes for a protein implicated in the synthesis of proteoglycans, concretely chondroitin sulphate. Chondroitin sulphate is not only a component of cartilage but is used in OA treatment, although its effectiveness is controversial.33 Another of the possible genes, TP63, is related with joint development and has been described as one of the loci determining facial morphology in humans.34 On its part, RUNX-2 is close to the CDC5L-SUPT3H locus, essential for both the differentiation of osteoblasts as well as skeletal morphogenesis. None of the previously mentioned loci have been studied further than their association. The only one that has been studied further is the locus with the FTO gene. This has been previously associated with obesity and has demonstrated a predisposition to weight increase through unclear factor that include regulation of food ingestion or energy production. This led to the suspicion that the association to OA was secondary to obesity because the latter is an important risk factor in knee OA. Upon analyzing this possibility it was seen that the association of OA and FTO disappeared after adjusting for BMI, suggesting a common genetic etiology and underlining the stratification by BMI in OA genetic studies.35
COL11A1Among the associations proposed for the first phase of the arcOGEN study and later unconfirmed, the COL11A1 gene is the most promising. This is due to the functional role of this gene that codifies for one of the 3 fibrils that form collagen typexi. This collagen binds to proteoglycan aggregates and anchors them to the extracellular matrix collagen network.36 It also stands out because some mutations in this gene leads to hereditary syndromes that include in their spectrum of manifestations, early onset OA.37 A more indirect evidence is its association with lumbar disc hernia (LDH) in Japanese patients, because the biology of the intervertebral disc and joint cartilage are related. The expression of COL11A1 is reduced in the intervertebral disc of patients with LDH and the LDH susceptibility gene (allele C) of the rs1676486 associated SNPs.38 However, rs1676486 is not associated to OA, although it is with the expression of COL11A1 in OA cartilage. In case this was not paradoxical enough, the COL11A1 most commonly associated SNPs with OA in the phase 1 of the arcOGEN study, rs2615977, shows no association with the expression of the gene in cartilage.39 It is possible that a recent meta-analysis of 9 GWAS of OA has led to clues that may explain these results, as there was an independent association of 2 SNPs of COL11A1 with hip OA localized in the gene limits.40 Therefore, it is possible that the paradoxes of previous studies are solved when considering the possibility of 2 independent associations, none of them coincident with those functionally studied until now.
DOT1L and Hip OAAs stated earlier, the association of DOT1L was discovered by studying an endophenotype of hip OA, the joint space width (JSW). Although it has been used as an example of the efficacy of endophenotypes, it should be noted that it is the only locus discovered in the GWAS that employed it, making it low yield for a study such as the Rotterdam cohort (6523 subjects) analyzing a quantitative variable (which normally provides more power than a dycotomic variable). Later, it has been shown that the same SNPs of DOT1L is associated with hip OA in males (P=7.8×10−9) in a study with a large sample size. DOT1L is a histone methyltranspherase that interacts with TCF-4 regulating the transcription of the Wnt pathway genes and influencing chondrogenic differentiation, expressed in the cartilage of patients with OA and its suppression inhibits the accumulation of proteoglycans and cartilage during chondrogenesis.14
MCF2L Helped by Genome Resequencing StudiesAnother recent analysis discovered the association of MCF2L with hip OA. This study used information from the 1000 genome project to obtain a large enough confidence level for this association. The 1000 genome project attempts to provide a complete catalog of all of the genetic variation in the human genome. Through last generation sequencing, followed by several stages of phenotyping and validation, common variants low frequency variants (0.5%–5%) and rare variants (<0.5%) are cataloged. This information was used to impute SNPs and rare variants that had not been genotyped in the phase 1 GWAS of arcOGEN. The comparison between cases and controls of more than 7 million variants with a frequency >1% led researchers to select 8 SNPs in 6 loci to genotype directly in the samples of the original GWAS. In this manner, new SNPs associated with MCF2L were found, whereas only rs11841874 had been documented before, making it a doubtful result and has not led to further study. Once the confidence level increased, this result was analyzed in other cohorts, leading to a significant GWAS level association (P meta-analysis=2×10−8). Little is known on the functional role of MCF2L other than it participates in the regulation of neurotrofin-3 that belongs to the NGF family, a proangiogenic factor whose expression in increased in OA chondrocytes.13
NCOA3 and Hip OAIn a hip OA GWAS meta-analysis, the association with rs6094710 (p=7.9×10−9) was discovered. This study gathered 4349 hip OA cases and 46 903 controls in the discovery phase and 11,277 cases and 67 473 controls in the replication, making it the largest up to date. The rs6094710 SNPs is close to the NCOA3, a gene with a reduced expression in OA cartilage. In addition, rs6094710 is in LD with a non-synonymous SNPs (Arg>Cys) that is probably harmful for a variant of the protein, leading to a possible molecular mechanism of action. NCOA3 might have a role in bone metabolism, being a coactivator of several nuclear receptors such as retinoids, vitamin D or thyroid hormone (T3). Although it might be likely that NCOA3 participates in chondrocyte mechanotransduction processes.15
DVWA and HLA of Class ii/iii and OA in Asian PatientsThe DVWA and HLA class ii/iii loci show a GWAS association that is restricted to the Asian population. These are 2 examples of the genetic heterogeneity existing between the European and Asian populations. The association of DVWA with knee OA was identified in a GWAS performed in the Japanese population.41 2 later specific studies have been performed on the same polymorphisms in the European population, but none of them showed an association.42,43 The function of this gene has no been widely studied. In the study that discovered its association, it was proven that DVWA interacts with ¿-tubulin differentially depending on the alleles of the 2 non synonymous SNPs that had shown an association. Later, there has been controversy on the nature of this gene. There is evidence that DVWA is really the 5′ portion of another gene, COL6A4.44 However, the recent cloning of the gene seems to have found another explanation, revealing 2 isoforms of the protein, a long one and a short one. The short one has no homology with COL6A4 and has specific expression in cartilage. The role of DVWA in chondrocyte differentiation and intracellular traffic has been hypothesized.45
The association of HLA was discovered in a GWAS performed in Japanese patients with knee OA. There were 2 SNPs associated: rs7775228 in HLA-DQ1B and rs10947262 in BTNL-2 (P=2.4×10−8 and P=6.7×10−8, respectively), localized in the HLA region of class ii/iii and which are not independent representing, therefore, a unique association.46 These SNPs are not associated with knee OA in the European samples of the same study or later ones.47,48 The importance of this association resides in that it might explain part of the inflammatory component of OA.
ALDH1A2 and Hand OAUp until recently, A2BP1 was considered hand OA's most relevant locus and rs716508 this phenotypes most strongly associated SNPs (P=1.81×10−5).49 However, recently discovered GWAS level associations have been described in hand OA.16 In the study's discovery phase, performed in Iceland, 55 associated variants were found (p<5×10−8). All of them are in LD in chromosome 15q22 and were classified into 2 groups considering the frequency of the at risk allele (41 and 52%) with 2 representative SNPs for each group, rs4238326 and rs3204689. Variants for both groups were tested in 5 additional European collections of samples, and the conjoined analysis showed a GWAS level of association for rs4238326 (OR=1.44; P=8.6×10−11) and rs3204689 (OR=1.46; P=1.1×10−11). In relation to the functional role of the locus, ALDH1A2 codifies for retinaldehide dehydrogenase 2, which catalizes the synthesis of retinoic acid, molecules which are relevant in the development of bone and cartilage.
The Role of Mitochondrial DNA in OAIn addition to nuclear genome, the possible role of haplogroups of DNAmt in OA has also been studied. Each mitochondrial haplogroup is defined by a particular combination of gene variants. Concretely, haplogroups J (m.4216T>C, m.10398A>G) and JT (m.4216T>C) were associated with a reduced risk of suffering knee OA and the J and J1c (m.14798T>C) groups were related to a reduced risk of hip OA in Spaniards.50,51 In the Asian population there was also an association with another 2 mitochondrial haplogroups, G and B/B4, which showed a susceptibility and protection effect, respectively.52 However, the arcOGEN study, with a larger sample size, (7393 cases and 5122 controls in the United Kingdom) did not find an association with any haplogroup of DNAmtmt.53 It has been suggested that differences in frequency in the haplogroups of different study populations (Spain and the UK) may justify the discrepancies, in addition to the lack of replication, which does not constitute enough evidence, such as that mentioned for loci GDF-5 and 7q22.54 Other associations have been found which may reinforce the implication of the mitochondrial haplogroups in the pathogenesis of OA, such as the association with serum collagen typeii and metalloproteinase levels.55
Future of OA Genetic StudiesThe study of genetic factors implicated in OA susceptibility is a field that has shown notable progress in recent years. There are currently 11 loci associated to OA in the European population at the level required for GWAS and 4 nearing this level. Even then, the number of susceptibility genes is relatively low if we compare it with other complex diseases. Field researchers have proposed a series of additional strategies to be applied in future studies that would help to complete the study of gene components in OA. Among these are increasing the sample size of studies, standardizing patient phenotypes and using the information of the 1000 genomes project to complete the genetic variation coverage. It may also be useful to perform other studies, such as genome resequenciation, or exome studies, in search of rare variants with a high penetrance and the study of microsatellites and VNTR, which are unstudied variants in GWAS. In addition, more information could be extracted from GWAS which have been already performed with new study strategies, as has occurred in meta-analysis of gene candidates or gene based analysis.
In relation to the search for rare variants, sequencing genes already associated may be of great interest, as they may come with rare variants such as has been seen in other diseases. However, one of these studies on GDF-5 found no variants contributing to OA susceptibility.56
With regard to microsatellites and VNTR, there are 2 that have shown an association with OA in some studies: a VNTR in ASPN (asporin) and a microsatellite in intron 1 of BMP-5. The association between the VNTR of ASPN as a genetic susceptibility factor for OA was detected in the Japanese population. The frequency of the 14 repetition allele of VNTR (D14) was high in knee OA patients compared to controls (P=0.000066). The D13 repetition had a lesser effect and in a contrary or protective direction, that is, elevated among controls.57 This association was not found in the European population, which demonstrates the existence of an ethnic component that differentiates both populations. On its part, the BMP-5 microsatellite, D6S1276, showed an association (P=0.018) with hip OA in female patients from the United Kingdom. Additionally, it was shown to affect the transcriptional activity of the gene in vitro. This finding led to further studies of BMP-5, demonstrating its association with another phenotype, knee OA in patients from 3 European countries.58,59 Because neither the association of ASPN nor that of BMP-5 have reached GWAS levels, their study has been limited, but suggests that variants in the number of copies may hide OA genetic factors.
Another strategy, as mentioned above, is extracting the information from an already performed GWAS. There is a lot of experience demonstrating the value of meta-analysis. Complementing this through the study of groups of particular genes for which there already is some type of independent evidence of their participation in OA would be useful. An approximation of this kind has been performed using evidence derived from the study of candidate genes. This analysis pointed out the association of hip OA and COL11A1 and VEGF.40 Another strategy that might be useful is to obtain statistically combined independent associations of the SNPs of a gene. This is what is called Gene-Based Analysis (GBA). It is based on the hypothesis that multiple genetic variants in disease-important genes might contribute, although none does it at a level strong enough to be detected individually. Some studies of this kind have been carried out in other diseases, without convincing results. It is possible that new tools are required to perform this type of analysis.
Therefore, even though the past years have led to many successes in the identification of loci associated with complex diseases, OA researchers are not satisfied, as we are aware that there are many more to identify. A whole series of approximations have been planned in order to help advancement in its study, although it is still unclear which will be more fruitful. It is possible that each one will provide some advances and little by little our knowledge of the genetic component of OA will be completed. Another aspect in which only the first steps have been walked until now is the identification of the causal variants of the increase in susceptibility and the mechanisms over which there is such an effect. This is an area that requires urgent development, as it is where there are more probabilities of contributing to the generation of knowledge and management of the disease.
Ethical ResponsibilitiesProtection of people and animalsThe authors declare that this study did not perform experiments on humans or animals.
Data confidentialityThe authors declare that they have followed the protocols of their workplace regarding the publication of data from patients, and all patients included in the study have received sufficient information and gave written informed consent to participate in the study.
Right to privacy and informed consentThe authors have obtained informed consent from patients and/or subjects referred to in the article. This document is in the possession of the corresponding author.
Conflicts of InterestThe authors declare no conflicts of interest.
Please cite this article as: Rodriguez-Fontenla C, Gonzalez A. Genética de la artrosis. Reumatol Clin. 2015;11:33–40.