To evaluate IP-10 gene expression in patients with SLE, and its possible relationship with disease activity.
Patients and methodsThis study included 120 patients diagnosed with SLE and 30 healthy controls. The relative gene expression of IP-10 was investigated with the Fold Change method, which was correlated with the level of lupus activity evaluated with the SLEDAI 2-K instrument.
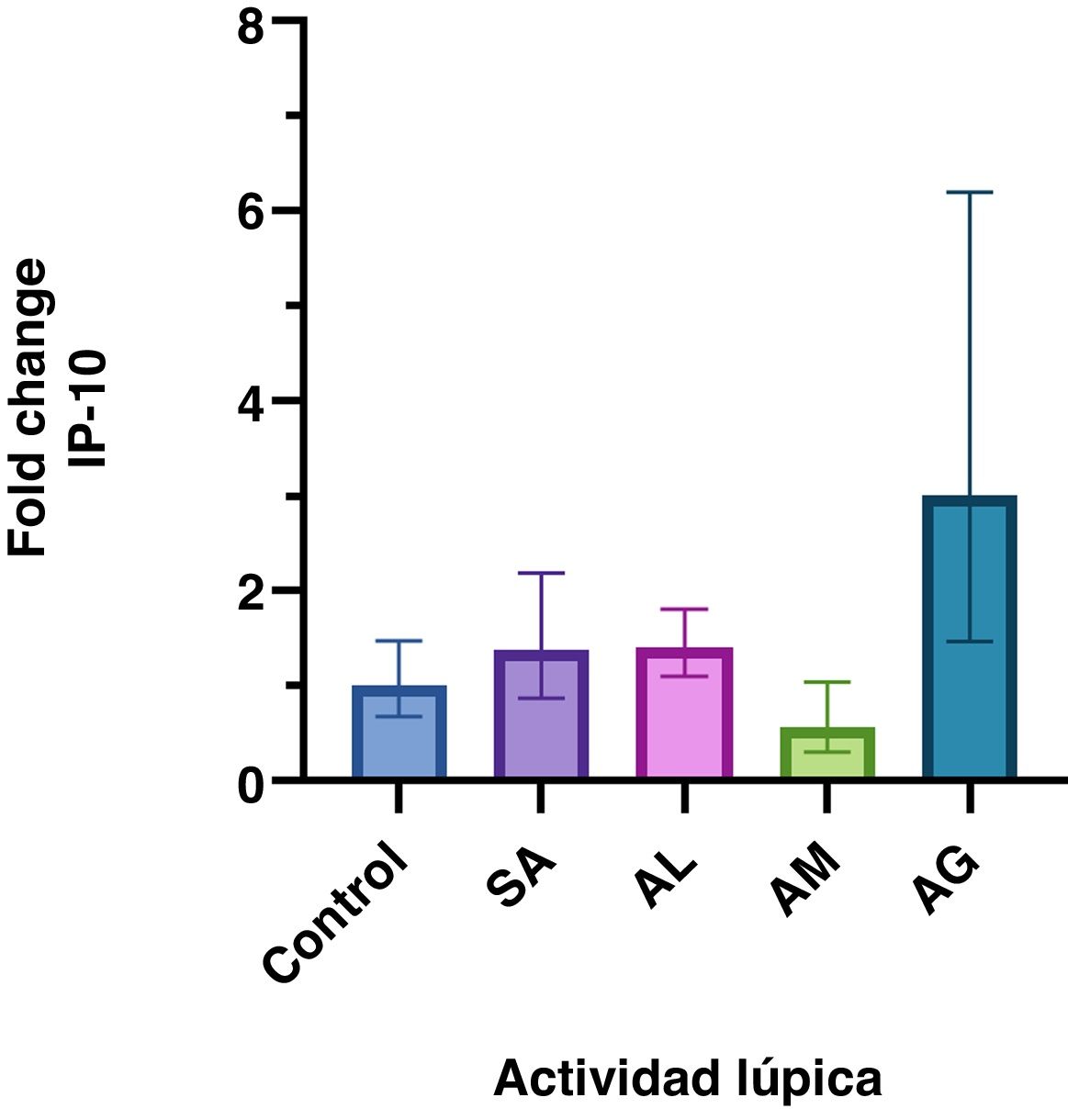
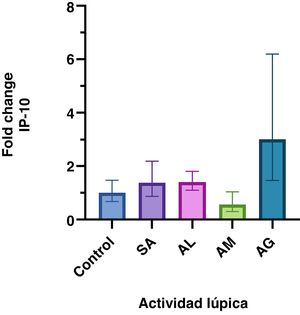
ResultsDifferent levels of gene expression were found according to the SLE activity (P = <.001). IP-10 gene expression levels were higher in patients with severe activity than in those with no activity, low activity, and moderate activity. The increase in gene expression in the severe activity group was significant with a Fold Change of 3
ConclusionThe significant increase in relative gene expression IP-10 may be a marker of severe lupus activity.
Evaluar la expresión génica del gen IP-10 en pacientes con LES, y su posible relación con la actividad de la enfermedad.
Pacientes y métodosEl estudio incluyó 120 pacientes diagnosticados con LES y 30 controles sanos. Se investigó la expresión génica relativa de IP-10 con el método Fold Change, la cual fue correlacionada con el nivel de actividad lúpica evaluado con el instrumento SLEDAI 2-K.
ResultadosSe encontró diferentes niveles en la expresión génica de IP-10 relacionada con la actividad lúpica (p= <0,001). Los niveles de expresión génica de IP-10 fueron mayores en los pacientes con actividad grave respecto a aquellos sin actividad, actividad baja y actividad moderada. El incremento en la expresión génica del grupo con actividad grave fue significativo con un Fold Change de 3.
ConclusiónEl incremento significativo en la expresión génica relativa IP-10 puede ser un marcador de actividad lúpica grave.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with heterogeneous presentation of symptoms, time-varying activity, multisystemic, characterised by the presence of antinuclear antibodies.1,2
Despite decades of studies, few biomarkers of SLE activity have been validated.3 The conventional markers (anti-C1q, anti-dsDNA, C3 and C4) are suboptimal, as they have a sensitivity of 50% and specificity of 75%.4 Hence the need for new biomarkers to determine response to treatment, and the need to modify treatment.
There is evidence that interferon (IFN) signalling is a major inducer in the pathophysiology of SLE. For example, the use of INF in other diseases induces SLE-associated antibodies. Therefore, overexpression of INF-stimulated genes in the blood of these patients has the potential to be a biomarker of activity.5
INF-induced chemokine protein 10 (IP-10 or CXCL10) is secreted by macrophages, monocytes, and endothelial cells in response to INF.6 Its CXCR3 receptor is expressed in Th1, B, NK lymphocytes and dendritic cells. Its functions include chemotactic activity, apoptosis induction, growth regulation, cell proliferation and angiogenesis. It is important because several studies have linked its serum levels to lupus activity and lupus nephritis.7
Although an increase in the relative expression of IP-10 has been found in leukocytes obtained from the peripheral blood of patients with SLE,8 there is little information correlating the degree of gene expression with disease activity, and therefore the aim of this study is to evaluate the relative gene expression of the IP-10 gene in people with SLE, and its possible relationship with disease activity.
Patients and methodsPatientsDuring the period between January 2018 and October 2019, 270 women who attended the rheumatology outpatient department or were admitted to the Central Military Hospital, classified as SLE cases according to the 1997 American College of Rheumatology criteria, were recruited. Their activity was quantified using the SLEDAI-2K instrument blinded to obtaining IP-10 gene expression,9 excluding those with active infectious processes over the last three months.
Thirty healthy women were also recruited for the control group who were not obese, and had no autoimmune diseases or active infections in the last three months. All the patients gave their informed consent, and this research study was approved by the bioethics and research committee of the Central Military Hospital.
Five groups were formed based on their SLEDAI-2K score, each with n = 30: 1.- control, 2.- no disease activity (NDA) score of zero to two, 3.- mild disease activity (MDA) score of three to four, 4.- moderate disease activity (ModDA) score of six to eight and 5.- severe disease activity (SDA) score of >eight.3
SamplesBlood samples were obtained by venous puncture from all the patients and processed in the molecular biology laboratory of the SEDENA Military School of Health Graduates, obtaining and quantifying total ribonucleic acid (RNA) from leukocytes in each of the samples.
In addition, primers were designed to amplify a specific region in exons 2 and 3 of the IP-10 gene and gene expression was measured using Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) of IP-10 messenger RNA (mRNA). The expression of an endogenous gene (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) was then obtained from all samples. KAPA SYBR FAST One-Step qRT-PCR Master Mix (2X), from Kappa Biosystem was the kit used.
Quantification of gene expressionThe relative gene expression of the groups was quantified by the fold change (FC) method, using the values of the cycles in which the samples begin to show significant fluorescence (Ct), obtained by qRT-PCR; Ct values are inversely proportional to the initial concentration of mRNA.
The first step in this method is to obtain the ΔCT which is the difference between the Ct means of the studied gene and the endogenous gene in a group, this value evaluates the expression of the gene in a group. Then, the ΔΔCT is obtained, which is the difference of the ΔCT between the groups with the disease and the control group, which evaluates gene behaviour in a disease. Finally, the FC is obtained using the formula 2−ΔΔCT and means the times a gene is over- or under-expressed on the logarithmic scale, in a group or groups with SLE with respect to the control group which has an FC value of 1.
Because of its continuous scale, tendency to normality and not representing a proportion, the overall statistical significance between groups is obtained with the ΔCT values by the analysis of variance test (ANOVA). Expression of the gene under study (IP-10) is considered to be significantly involved in disease activity when the FC of the group is greater than two.
GraphPad Prism 8.4.0 software was used for the statistical analysis.
ResultsNot normally distributed data are presented as medians (IQR). Of all the patients included, the median age was 38 years (29, 53). The women with SLE had a median SLEDAI-2K score of eight (four, 27) and age of 42 years (34, 54). The characteristics by group and FC analysis are shown in Table 1 and Fig. 1 plots the FC of the groups. The difference in the relative expression of IP-10 between the groups was statistically significant (F = 5.3 [4104.8], P = <.001) determined by one-way ANOVA (Brown-Forsythe).
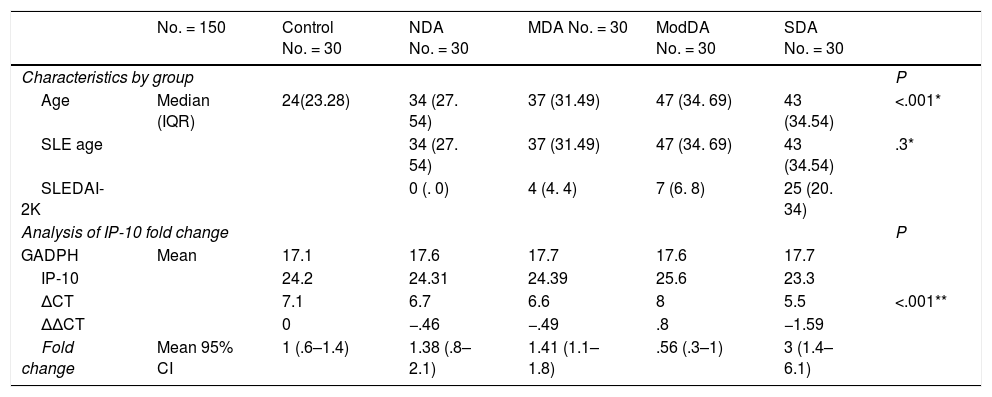
Characteristics of the groups and relative gene expression (fold change) of IP-10.
| No. = 150 | Control No. = 30 | NDA No. = 30 | MDA No. = 30 | ModDA No. = 30 | SDA No. = 30 | ||
|---|---|---|---|---|---|---|---|
| Characteristics by group | P | ||||||
| Age | Median (IQR) | 24(23.28) | 34 (27. 54) | 37 (31.49) | 47 (34. 69) | 43 (34.54) | <.001* |
| SLE age | 34 (27. 54) | 37 (31.49) | 47 (34. 69) | 43 (34.54) | .3* | ||
| SLEDAI-2K | 0 (. 0) | 4 (4. 4) | 7 (6. 8) | 25 (20. 34) | |||
| Analysis of IP-10 fold change | P | ||||||
| GADPH | Mean | 17.1 | 17.6 | 17.7 | 17.6 | 17.7 | |
| IP-10 | 24.2 | 24.31 | 24.39 | 25.6 | 23.3 | ||
| ΔCT | 7.1 | 6.7 | 6.6 | 8 | 5.5 | <.001** | |
| ΔΔCT | 0 | −.46 | −.49 | .8 | −1.59 | ||
| Fold change | Mean 95% CI | 1 (.6–1.4) | 1.38 (.8–2.1) | 1.41 (1.1–1.8) | .56 (.3–1) | 3 (1.4–6.1) | |
SDA: Severe disease activity; MDA: Mild disease activity; ModDA: Moderate activity; SLE age: Age of patients with systemic lupus erythematosus; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; CI: Confidence interval; IQR: Interquartile range; IP-10: Interferon gamma-induced protein 10; NDA: No disease activity.
Relative IP-10 gene expression according to SLE activity. The geometric mean and 95% confidence interval of the IP-10 fold change in each group are presented. Only the group with severe disease activity shows a significant increase in relative gene expression.
LES: systemic lupus erythematosus; MDA: Mild disease activity; ModDA: Moderate disease activity; NDA: No disease activity; SDA: Severe disease activity.
This study demonstrated a significant increase (FC > 2) in IP-10 gene expression in the patients with SLE and SDA, who had a greater increase in IP-10 expression than the other subjects. Apart from the MDA group, the FC values were higher in those diagnosed with SLE.
Although there was a statistically significant difference between the age of all the groups, when only patients diagnosed with SLE were considered, the P-value was .3. Therefore, the variation in gene expression observed in this study cannot be considered to be associated with age.
The findings in the ModDA group could be explained by differences in response to treatment, as it has been found that IP-10 expression is reduced in patients who respond well to immunomodulatory treatment. It is possible that the ModDA group represents the population with a good response, patients with MDA or SDA, those starting on therapy or newly diagnosed and those with SDA that is resistant to treatment.10
There is also evidence that the predominance of the Th1 phenotype in CD4+ lymphocytes in SLE patients is associated with increased lupus activity. INF secreted by Th1 lymphocytes promotes the synthesis and release of INF response products such as IP-10. By contrast, interleukins secreted by Th2 lymphocytes antagonise the secretion and action of mediators released by Th1 lymphocytes, including INF.11 Therefore, the lower IP-10 gene expression in the ModDA group may be due to a predominance of Th2 lymphocytes, and the SDA group has higher IP-10 gene expression due to a predominance of Th1 lymphocytes.
In conclusion, it could be suggested that the significant increase in IP-10 gene expression may be a biomarker of severe lupus activity, which implies that it could be used to identify patients that require adjustment of treatment and stricter surveillance. As this was a preliminary study, the patients’ treatments and CD4+ lymphocyte phenotype were not collected and further prospective studies with larger case numbers that consider these variables are needed to corroborate our observations.
FundingNo funding was received for this paper.
Conflict of interestsThe authors have no conflict of interests to declare.
Please cite this article as: Torres-Vázquez J, Uriel Vázquez-Medina M, Comoto-Santacruz DA, Pradillo-Macias ME, Muñoz-Monroy OE, Martínez-Cuazitl A. Relación de la expresión génica de IP-10 con la actividad del lupus eritematoso sistémico. Reumatol Clin. 2022;18:91–93.